Gene
KWMTBOMO02773
Pre Gene Modal
BGIBMGA003492
Annotation
PREDICTED:_F-box/LRR-repeat_protein_14-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.882
Sequence
CDS
ATGGAACTTCCCTATGAGATTTTAGTGTATTTATTAAGATTTCTCCCAAAATCTGATCGAAAATCAGCGAGTGAAACATGTCGTTCGTGGTATATAGCAGCAAATGATCAACATTTTCTGAAAAACAAAGTAGTTATATTTTATAAATCAGTAATTAATGATGAAAACTCATCACCAATCCAAATATTTGAAAATTCTTTTACATCATACCATAACTTTTTTTTCAACGAAGTCGAATTATCAAGCAAGTTAAATGATTTTTGGATCAAAATAGGAGAAAACATAAAATGTTTATCTCTGACAAGCTGTGATGTAAATGAAAAGGTATTTGTTCATATTTTACAAAATTGTACTAATTTAAAAGAGCTGCACATTAAGAGTTGCAAAGAACTATTTATGTCTGGTCGATTGCTTGAAGGAAAAACAGAAGGTCTTTTAGTTACTAAACTTGAACATTTGAAATGTCTCAGCTTATCTGGAAATCAGTACTTAACTGATGCTTTATTTAATAGATTTATATCAGCATCTCCTGGATTAGAGGAACTTAATCTATCTGGATGTTCTCTACAATTTCATCTAGGTCTTGTCAAAAAGTTCTATCCAAATGGAACAGACATTTTTCAGAATCCATCAGAGAGTGTTTTAACTTTTCATTTTATTCATCAAATGATAATCTCCAGAGCTGACAAAATAAAACGACTGTTGTTTTCTAATACACTAATTGATGGTGCTGCTCTTAAAACTTTATCAGAGATAAAAAATCTTCAATTAGAATCACTGGAAGTGCATGCTTGTGATCAACTCACTAATACAGGCATTTTATCACTTACAAGCCACCAGATCTTCTTAAAGGAACTTGATATTGGACTGTGTACTAGAGTTACAGATCAGTCCCTTGTATATATTTGTAAAAATCTAGTCAACTTAGAAAACCTCAATATTCAAAGATGTCGAGCTGTGACTGATTTAGGTATTGCAGAGTTGGGTAAATTAATTAAATTAAAATCATTAAACATCTCTCGGTGCGAACTGTTGACTAAGGATGGATTATGTAAAGGTATATGTGCTGAAGTAAATGTGGTATTAGAAGATTTAGATATACATTCTCTAAATTTAGATCAAACAGGCCTCATAATGATCTCTGAAAAATTACCAAACTTACGTGCATTAGACTTGAGTTATTGTTTTAATGCAGTTACTGATACATCAATCCAAGTAATATTTAAGAATCAAGCATTTCTTAAAGTATTAAAAATTAGCCACTGTGATAAAGTCTCTGATGCTGGTTTAACTGGTATGGGGAAAATAGAAACTGAAACAAATGATGATGGACCGATAGAATCCAATTATAATACAGAACCAACATTACCAAGGATATATTTAGGATCCAGAGCCGAAGAAGAGATTGTTCGTGATGCCAAACGCAAGCGTGATGTAATGCGTATGTGTGAGAAGATATCAACAAATTCCTATACGGGCTATTCACTTGCAAGGCTCAAGGGTCTGCGTGATTTGGATTTGAGTGGATGCAACAGAATTACAGATGTTAGCCTTACTTATGCCTTTAATTTTAAAGAACTTATCAATTTGAACATTAGCAGATGTCAACAAATAACACACCAAGGTGTTGAGCATTTAGTTAAGAATTGTCAGAGTATTGAATATATCAATCTCAATGACTGCTATAACTTAAATGATCATGCTGTCAAAGAAATTGTTCAACAATTGAAAAGGTTAAAACAACTGGAATTAAGGGGATGCAATCAATTAACAGACAAGACACTGGAATATGTCCGATCTTATTGCGAGAAATTAAAATTTTTGGATGTACAAGGCTGCAGGAATATGTCACCAGAACTTGCTTGCTCTATTGGAAGTTTACCTACCCTCCACACTGTGTTGATGACCAAACCTGGACCATATGTAACAGATGATGTAAAGCAACGTTCGCCTGCACCAGCATTCCTTCCTGCCTTAATGAGAAAACTTCGTTTACACTAA
Protein
MELPYEILVYLLRFLPKSDRKSASETCRSWYIAANDQHFLKNKVVIFYKSVINDENSSPIQIFENSFTSYHNFFFNEVELSSKLNDFWIKIGENIKCLSLTSCDVNEKVFVHILQNCTNLKELHIKSCKELFMSGRLLEGKTEGLLVTKLEHLKCLSLSGNQYLTDALFNRFISASPGLEELNLSGCSLQFHLGLVKKFYPNGTDIFQNPSESVLTFHFIHQMIISRADKIKRLLFSNTLIDGAALKTLSEIKNLQLESLEVHACDQLTNTGILSLTSHQIFLKELDIGLCTRVTDQSLVYICKNLVNLENLNIQRCRAVTDLGIAELGKLIKLKSLNISRCELLTKDGLCKGICAEVNVVLEDLDIHSLNLDQTGLIMISEKLPNLRALDLSYCFNAVTDTSIQVIFKNQAFLKVLKISHCDKVSDAGLTGMGKIETETNDDGPIESNYNTEPTLPRIYLGSRAEEEIVRDAKRKRDVMRMCEKISTNSYTGYSLARLKGLRDLDLSGCNRITDVSLTYAFNFKELINLNISRCQQITHQGVEHLVKNCQSIEYINLNDCYNLNDHAVKEIVQQLKRLKQLELRGCNQLTDKTLEYVRSYCEKLKFLDVQGCRNMSPELACSIGSLPTLHTVLMTKPGPYVTDDVKQRSPAPAFLPALMRKLRLH
Summary
Uniprot
H9J1V5
A0A2H1WLY3
A0A2A4JY87
A0A194PNJ8
A0A2A4JX94
A0A194QRK0
+ More
A0A1E1W7Q5 A0A0L7LDU3 A0A1E1WP54 A0A212ETB3 A0A067QMJ0 A0A1B6GGS5 A0A182GEP0 A0A1B6KZ31 A0A2J7PT45 A0A1Q3FK81 Q0IEJ5 A0A2A3ECU9 A0A0N0U6Q5 A0A154P7Y1 A0A088AM26 A0A0L7RHY2 B0WMK4 A0A2P8Y5G4 A0A1B6CRF7 A0A336KDX7 A0A1J1J2F0 A0A182QHX4 A0A336M0E8 A0A182WUD3 A0A182V0V6 A0A0E4GBG0 A0A182TWV0 A0A182HW42 A0A182NSX3 A0A182RD64 A0A182MTK7 A0A182KKT9 A0A182JYE3 A0A182YAV0 A0A182WDR9 A0A182P5C6 A0A084WLU5 A0A182SZW0 A0A182FTK9 W5JG98 A0A2M4AGU7 A0A182ILJ2 A0A2M4AH07 A0A2M4AGY4 A0A1I8PYR9 A0A0K8TLT6 A0A1A9W9U3 A0A0L0BSQ7 A0A067QKD9 B3MFE0 A0A1I8ME70 A0A3B0J5T7 A0A1A9VXJ0 B4MK52 W8C7V8 A0A0A1WSN2 A0A1B0A7F0 A0A1B0G1T5 A0A1W4UXD5 A0A1A9Y667 A0A034WUL1 B4HSA1 A0A0K8VKI9 Q7K0V7 A0A1B0BLM6 A0A0B4KEH1 B4JVD8 B4P3D0 B4GGJ3 B3N857 Q28Y47 A0A0P4VX65 B4QG95 A0A0K8TV16 B4LN93 B4KTR5 T1GNE4 A0A2M4AKF9 A0A2M4AKF4 A0A2M4BJ29 A0A2M4BJ44 A0A1W4WT00 A0A084WLU3 A0A0A1WKF8 E9G608 B0WW37 A0A1S4HDZ8 Q16NN3 A0A1S4FUY3 A0A182R028 A0A2R5LAG0 S7PZM5 A0A182MTJ2
A0A1E1W7Q5 A0A0L7LDU3 A0A1E1WP54 A0A212ETB3 A0A067QMJ0 A0A1B6GGS5 A0A182GEP0 A0A1B6KZ31 A0A2J7PT45 A0A1Q3FK81 Q0IEJ5 A0A2A3ECU9 A0A0N0U6Q5 A0A154P7Y1 A0A088AM26 A0A0L7RHY2 B0WMK4 A0A2P8Y5G4 A0A1B6CRF7 A0A336KDX7 A0A1J1J2F0 A0A182QHX4 A0A336M0E8 A0A182WUD3 A0A182V0V6 A0A0E4GBG0 A0A182TWV0 A0A182HW42 A0A182NSX3 A0A182RD64 A0A182MTK7 A0A182KKT9 A0A182JYE3 A0A182YAV0 A0A182WDR9 A0A182P5C6 A0A084WLU5 A0A182SZW0 A0A182FTK9 W5JG98 A0A2M4AGU7 A0A182ILJ2 A0A2M4AH07 A0A2M4AGY4 A0A1I8PYR9 A0A0K8TLT6 A0A1A9W9U3 A0A0L0BSQ7 A0A067QKD9 B3MFE0 A0A1I8ME70 A0A3B0J5T7 A0A1A9VXJ0 B4MK52 W8C7V8 A0A0A1WSN2 A0A1B0A7F0 A0A1B0G1T5 A0A1W4UXD5 A0A1A9Y667 A0A034WUL1 B4HSA1 A0A0K8VKI9 Q7K0V7 A0A1B0BLM6 A0A0B4KEH1 B4JVD8 B4P3D0 B4GGJ3 B3N857 Q28Y47 A0A0P4VX65 B4QG95 A0A0K8TV16 B4LN93 B4KTR5 T1GNE4 A0A2M4AKF9 A0A2M4AKF4 A0A2M4BJ29 A0A2M4BJ44 A0A1W4WT00 A0A084WLU3 A0A0A1WKF8 E9G608 B0WW37 A0A1S4HDZ8 Q16NN3 A0A1S4FUY3 A0A182R028 A0A2R5LAG0 S7PZM5 A0A182MTJ2
Pubmed
19121390
26354079
26227816
22118469
24845553
26483478
+ More
17510324 29403074 12364791 20966253 25244985 24438588 20920257 23761445 26369729 26108605 17994087 25315136 24495485 25830018 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 21292972
17510324 29403074 12364791 20966253 25244985 24438588 20920257 23761445 26369729 26108605 17994087 25315136 24495485 25830018 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 21292972
EMBL
BABH01007931
ODYU01009501
SOQ53986.1
NWSH01000406
PCG76638.1
KQ459597
+ More
KPI94892.1 PCG76635.1 KQ461181 KPJ07605.1 GDQN01008077 JAT82977.1 JTDY01001546 KOB73575.1 GDQN01002357 JAT88697.1 AGBW02012631 OWR44694.1 KK853244 KDR09426.1 GECZ01008149 JAS61620.1 JXUM01011720 KQ560337 KXJ82977.1 GEBQ01023258 GEBQ01013810 JAT16719.1 JAT26167.1 NEVH01021921 PNF19510.1 GFDL01007131 JAV27914.1 CH477655 EAT37715.1 KZ288282 PBC29557.1 KQ435726 KOX78251.1 KQ434826 KZC07444.1 KQ414584 KOC70602.1 DS231998 EDS31075.1 PYGN01000905 PSN39498.1 GEDC01021515 JAS15783.1 UFQS01000223 UFQT01000223 SSX01615.1 SSX21995.1 CVRI01000067 CRL06520.1 AXCN02002134 UFQT01000218 SSX21897.1 AAAB01008816 HACL01000130 CFW94424.1 APCN01001462 AXCM01003024 ATLV01024291 KE525351 KFB51189.1 ADMH02001295 ETN63111.1 GGFK01006517 MBW39838.1 GGFK01006749 MBW40070.1 GGFK01006734 MBW40055.1 GDAI01002703 JAI14900.1 JRES01001421 KNC23051.1 KK853333 KDR08366.1 CH902619 EDV35614.2 OUUW01000001 SPP75203.1 CH963846 EDW72491.2 GAMC01007806 JAB98749.1 GBXI01012480 JAD01812.1 CCAG010006899 GAKP01000628 JAC58324.1 CH480816 EDW46996.1 GDHF01012918 JAI39396.1 AE013599 AY069552 AAF59035.1 AAL39697.1 JXJN01016484 AGB93345.1 CH916375 EDV98406.1 CM000157 EDW89403.2 CH479183 EDW35613.1 CH954177 EDV59470.2 CM000071 EAL26118.2 GDRN01106551 JAI57551.1 CM000362 CM002911 EDX06245.1 KMY92354.1 GDHF01034000 JAI18314.1 CH940648 EDW60097.2 CH933808 EDW09648.2 CAQQ02141887 CAQQ02141888 CAQQ02141889 CAQQ02141890 GGFK01007949 MBW41270.1 GGFK01007948 MBW41269.1 GGFJ01003935 MBW53076.1 GGFJ01003934 MBW53075.1 ATLV01024290 KFB51187.1 GBXI01015272 JAC99019.1 GL732533 EFX84856.1 DS232137 EDS35869.1 AAAB01008986 CH477816 EAT35952.1 AXCN02001087 GGLE01002309 MBY06435.1 KE163836 EPQ14042.1 AXCM01001233
KPI94892.1 PCG76635.1 KQ461181 KPJ07605.1 GDQN01008077 JAT82977.1 JTDY01001546 KOB73575.1 GDQN01002357 JAT88697.1 AGBW02012631 OWR44694.1 KK853244 KDR09426.1 GECZ01008149 JAS61620.1 JXUM01011720 KQ560337 KXJ82977.1 GEBQ01023258 GEBQ01013810 JAT16719.1 JAT26167.1 NEVH01021921 PNF19510.1 GFDL01007131 JAV27914.1 CH477655 EAT37715.1 KZ288282 PBC29557.1 KQ435726 KOX78251.1 KQ434826 KZC07444.1 KQ414584 KOC70602.1 DS231998 EDS31075.1 PYGN01000905 PSN39498.1 GEDC01021515 JAS15783.1 UFQS01000223 UFQT01000223 SSX01615.1 SSX21995.1 CVRI01000067 CRL06520.1 AXCN02002134 UFQT01000218 SSX21897.1 AAAB01008816 HACL01000130 CFW94424.1 APCN01001462 AXCM01003024 ATLV01024291 KE525351 KFB51189.1 ADMH02001295 ETN63111.1 GGFK01006517 MBW39838.1 GGFK01006749 MBW40070.1 GGFK01006734 MBW40055.1 GDAI01002703 JAI14900.1 JRES01001421 KNC23051.1 KK853333 KDR08366.1 CH902619 EDV35614.2 OUUW01000001 SPP75203.1 CH963846 EDW72491.2 GAMC01007806 JAB98749.1 GBXI01012480 JAD01812.1 CCAG010006899 GAKP01000628 JAC58324.1 CH480816 EDW46996.1 GDHF01012918 JAI39396.1 AE013599 AY069552 AAF59035.1 AAL39697.1 JXJN01016484 AGB93345.1 CH916375 EDV98406.1 CM000157 EDW89403.2 CH479183 EDW35613.1 CH954177 EDV59470.2 CM000071 EAL26118.2 GDRN01106551 JAI57551.1 CM000362 CM002911 EDX06245.1 KMY92354.1 GDHF01034000 JAI18314.1 CH940648 EDW60097.2 CH933808 EDW09648.2 CAQQ02141887 CAQQ02141888 CAQQ02141889 CAQQ02141890 GGFK01007949 MBW41270.1 GGFK01007948 MBW41269.1 GGFJ01003935 MBW53076.1 GGFJ01003934 MBW53075.1 ATLV01024290 KFB51187.1 GBXI01015272 JAC99019.1 GL732533 EFX84856.1 DS232137 EDS35869.1 AAAB01008986 CH477816 EAT35952.1 AXCN02001087 GGLE01002309 MBY06435.1 KE163836 EPQ14042.1 AXCM01001233
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000027135 UP000069940 UP000249989 UP000235965 UP000008820 UP000242457 UP000053105 UP000076502 UP000005203 UP000053825 UP000002320 UP000245037 UP000183832 UP000075886 UP000076407 UP000075903 UP000075902 UP000075840 UP000075884 UP000075900 UP000075883 UP000075882 UP000075881 UP000076408 UP000075920 UP000075885 UP000030765 UP000075901 UP000069272 UP000000673 UP000075880 UP000095300 UP000091820 UP000037069 UP000007801 UP000095301 UP000268350 UP000078200 UP000007798 UP000092445 UP000092444 UP000192221 UP000092443 UP000001292 UP000000803 UP000092460 UP000001070 UP000002282 UP000008744 UP000008711 UP000001819 UP000000304 UP000008792 UP000009192 UP000015102 UP000192223 UP000000305
UP000027135 UP000069940 UP000249989 UP000235965 UP000008820 UP000242457 UP000053105 UP000076502 UP000005203 UP000053825 UP000002320 UP000245037 UP000183832 UP000075886 UP000076407 UP000075903 UP000075902 UP000075840 UP000075884 UP000075900 UP000075883 UP000075882 UP000075881 UP000076408 UP000075920 UP000075885 UP000030765 UP000075901 UP000069272 UP000000673 UP000075880 UP000095300 UP000091820 UP000037069 UP000007801 UP000095301 UP000268350 UP000078200 UP000007798 UP000092445 UP000092444 UP000192221 UP000092443 UP000001292 UP000000803 UP000092460 UP000001070 UP000002282 UP000008744 UP000008711 UP000001819 UP000000304 UP000008792 UP000009192 UP000015102 UP000192223 UP000000305
Interpro
Gene 3D
ProteinModelPortal
H9J1V5
A0A2H1WLY3
A0A2A4JY87
A0A194PNJ8
A0A2A4JX94
A0A194QRK0
+ More
A0A1E1W7Q5 A0A0L7LDU3 A0A1E1WP54 A0A212ETB3 A0A067QMJ0 A0A1B6GGS5 A0A182GEP0 A0A1B6KZ31 A0A2J7PT45 A0A1Q3FK81 Q0IEJ5 A0A2A3ECU9 A0A0N0U6Q5 A0A154P7Y1 A0A088AM26 A0A0L7RHY2 B0WMK4 A0A2P8Y5G4 A0A1B6CRF7 A0A336KDX7 A0A1J1J2F0 A0A182QHX4 A0A336M0E8 A0A182WUD3 A0A182V0V6 A0A0E4GBG0 A0A182TWV0 A0A182HW42 A0A182NSX3 A0A182RD64 A0A182MTK7 A0A182KKT9 A0A182JYE3 A0A182YAV0 A0A182WDR9 A0A182P5C6 A0A084WLU5 A0A182SZW0 A0A182FTK9 W5JG98 A0A2M4AGU7 A0A182ILJ2 A0A2M4AH07 A0A2M4AGY4 A0A1I8PYR9 A0A0K8TLT6 A0A1A9W9U3 A0A0L0BSQ7 A0A067QKD9 B3MFE0 A0A1I8ME70 A0A3B0J5T7 A0A1A9VXJ0 B4MK52 W8C7V8 A0A0A1WSN2 A0A1B0A7F0 A0A1B0G1T5 A0A1W4UXD5 A0A1A9Y667 A0A034WUL1 B4HSA1 A0A0K8VKI9 Q7K0V7 A0A1B0BLM6 A0A0B4KEH1 B4JVD8 B4P3D0 B4GGJ3 B3N857 Q28Y47 A0A0P4VX65 B4QG95 A0A0K8TV16 B4LN93 B4KTR5 T1GNE4 A0A2M4AKF9 A0A2M4AKF4 A0A2M4BJ29 A0A2M4BJ44 A0A1W4WT00 A0A084WLU3 A0A0A1WKF8 E9G608 B0WW37 A0A1S4HDZ8 Q16NN3 A0A1S4FUY3 A0A182R028 A0A2R5LAG0 S7PZM5 A0A182MTJ2
A0A1E1W7Q5 A0A0L7LDU3 A0A1E1WP54 A0A212ETB3 A0A067QMJ0 A0A1B6GGS5 A0A182GEP0 A0A1B6KZ31 A0A2J7PT45 A0A1Q3FK81 Q0IEJ5 A0A2A3ECU9 A0A0N0U6Q5 A0A154P7Y1 A0A088AM26 A0A0L7RHY2 B0WMK4 A0A2P8Y5G4 A0A1B6CRF7 A0A336KDX7 A0A1J1J2F0 A0A182QHX4 A0A336M0E8 A0A182WUD3 A0A182V0V6 A0A0E4GBG0 A0A182TWV0 A0A182HW42 A0A182NSX3 A0A182RD64 A0A182MTK7 A0A182KKT9 A0A182JYE3 A0A182YAV0 A0A182WDR9 A0A182P5C6 A0A084WLU5 A0A182SZW0 A0A182FTK9 W5JG98 A0A2M4AGU7 A0A182ILJ2 A0A2M4AH07 A0A2M4AGY4 A0A1I8PYR9 A0A0K8TLT6 A0A1A9W9U3 A0A0L0BSQ7 A0A067QKD9 B3MFE0 A0A1I8ME70 A0A3B0J5T7 A0A1A9VXJ0 B4MK52 W8C7V8 A0A0A1WSN2 A0A1B0A7F0 A0A1B0G1T5 A0A1W4UXD5 A0A1A9Y667 A0A034WUL1 B4HSA1 A0A0K8VKI9 Q7K0V7 A0A1B0BLM6 A0A0B4KEH1 B4JVD8 B4P3D0 B4GGJ3 B3N857 Q28Y47 A0A0P4VX65 B4QG95 A0A0K8TV16 B4LN93 B4KTR5 T1GNE4 A0A2M4AKF9 A0A2M4AKF4 A0A2M4BJ29 A0A2M4BJ44 A0A1W4WT00 A0A084WLU3 A0A0A1WKF8 E9G608 B0WW37 A0A1S4HDZ8 Q16NN3 A0A1S4FUY3 A0A182R028 A0A2R5LAG0 S7PZM5 A0A182MTJ2
PDB
4R5C
E-value=0.00245825,
Score=99
Ontologies
KEGG
GO
PANTHER
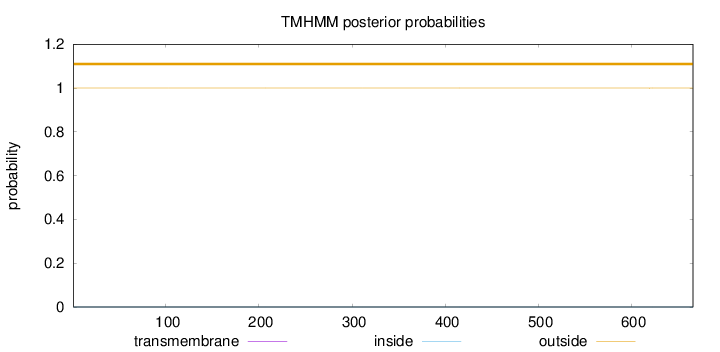
Topology
Length:
666
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00144
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00010
outside
1 - 666
Population Genetic Test Statistics
Pi
282.340759
Theta
207.313187
Tajima's D
0.941676
CLR
27.4132
CSRT
0.637818109094545
Interpretation
Uncertain
