Gene
KWMTBOMO02771
Pre Gene Modal
BGIBMGA003491
Annotation
PREDICTED:_serine_proteinase_stubble-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.337 PlasmaMembrane Reliability : 1.61
Sequence
CDS
ATGTGTCTGTGGCTGTTGACAGCTTGTTTTGGGCTATTGCCTGCAAACCCGTCCCCGATTGTGATAGTTCCTGGTGTGCCCGCGCCTATAATAGTGCCTACAACTGCAGCCACGACAGCAGCCACAACGACTCAGGCGCCAACAGCACCTCCGACGATGCCTGTAAATATAACCACTTCAGTACCGCCGACTATGCCGTCAACTATGCCGTCGACAATGCCGTCAACTATGCCGTCGACAATGCCGTCATCTATGCCGTCGACAATGCCGTCAACTATGCCGACGACAATGCCGCCAACCATGCCTCCTACTATGCCACCTACAATGCCTTCGGTTACTACAGCTCCACCGACGACTACTACTACTACTGCAGCTCCTACTCCGCCTCCGACAACAGCAGCTCCCACAACTTCAGGTCGATTGTTATTACCTGTTATATTTTGTAATGACCCTGATGTGGTGTGTTTATTAAATCCTGATGAAATCGCAGGGCCTCTACCGATCGATCCCAGGCTTGGAACTACTCCGGGCCCTCTGGCAGCTCTGCAAATACAGCCAATGGGTTATAGTTCTCAAGTACCATCTTACGATGCCGCTGTAAAGTTTCCAAGACAAAAAAGAAGTATGAGACCGCTTAATGTGAAAAACTTTTTGAGCAACTTGTCGAAGCCTGATGAACATCACGTAATTCACAAAAGGCAAAGTAACAACTGCAGATGTGTAACCGCTGGAACATGTATTCGATTTCCCATGATCGATGTCAGAATTGTTACTCCGGCAGTAAGTCAATGTCTAACAGGACAAGAATATTGTTGTGGTAATACAACTATGAACATGACGCAGATAGGATGTGGAACCCTACAATCGGTACCAACTACAGGAGTTACACCTATGGCTGGTCAAGCTAATTTCGGTGAATTTCCTTGGCAGGCCCTTATTTTAACGAAACAAAACGAGTACATCGGCGGAGGAGTTCTTATTGATAATATGAACGTTCTAACCGTTACTCATAGGATCATGTCTTATGTGGTATCAGGAACAGCTCCGAATGTGAAGGTTCGTCTCGGTGAGTGGGACGCCGCTGGCACGTATGAACCGGTCGCCCACCAAGAATACACAATACAAAGAGTGTCAAGTCACCCATCCTACAACGCTAACACGCTTCAGTTTGACGTCACAGTGCTTCGATTGGCAAGCCCGGTTCCGTTTACACCGACAACAGGAGCTGCAACTACTATCAACAGAGCGTGTCTTCCTTCAGCGGCTAGTGCCACCTTTACAGGTCAAAGATGCATGGTCTCCGGCTGGGGCAAGGATGCATTCGGTGCTCAAGGACAATTTCAAAATATTCTGAAAAAGGTCGATGTCCCAATTGTTGCTCCAGACGTTTGTCAAACGCAACTGAGAACTGCTCGGCTCGGGCCCACTTATGTTCTGGATACTACGTCATTCGTATGTGCCGGCGGAGAGCCCAATAAGGATGCTTGCACGGGCGATGGAGGTTCCGGTCTTGTGTGTCAAACAAATGGCCGTTGGATGGTGGTCGGGCTGGTCGCATGGGGTCTAGGTTGTGCCAACGCCAATGTGTCCGCTGTGTACGTCAATGTCGCGGGTTTCTTACCTTGGATACAACAACAAGTTGCCGCTGCTTAA
Protein
MCLWLLTACFGLLPANPSPIVIVPGVPAPIIVPTTAATTAATTTQAPTAPPTMPVNITTSVPPTMPSTMPSTMPSTMPSTMPSSMPSTMPSTMPTTMPPTMPPTMPPTMPSVTTAPPTTTTTTAAPTPPPTTAAPTTSGRLLLPVIFCNDPDVVCLLNPDEIAGPLPIDPRLGTTPGPLAALQIQPMGYSSQVPSYDAAVKFPRQKRSMRPLNVKNFLSNLSKPDEHHVIHKRQSNNCRCVTAGTCIRFPMIDVRIVTPAVSQCLTGQEYCCGNTTMNMTQIGCGTLQSVPTTGVTPMAGQANFGEFPWQALILTKQNEYIGGGVLIDNMNVLTVTHRIMSYVVSGTAPNVKVRLGEWDAAGTYEPVAHQEYTIQRVSSHPSYNANTLQFDVTVLRLASPVPFTPTTGAATTINRACLPSAASATFTGQRCMVSGWGKDAFGAQGQFQNILKKVDVPIVAPDVCQTQLRTARLGPTYVLDTTSFVCAGGEPNKDACTGDGGSGLVCQTNGRWMVVGLVAWGLGCANANVSAVYVNVAGFLPWIQQQVAAA
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
2B9L
E-value=7.06792e-43,
Score=439
Ontologies
GO
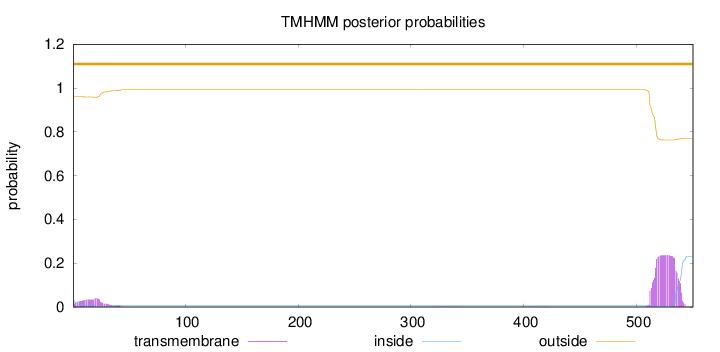
Topology
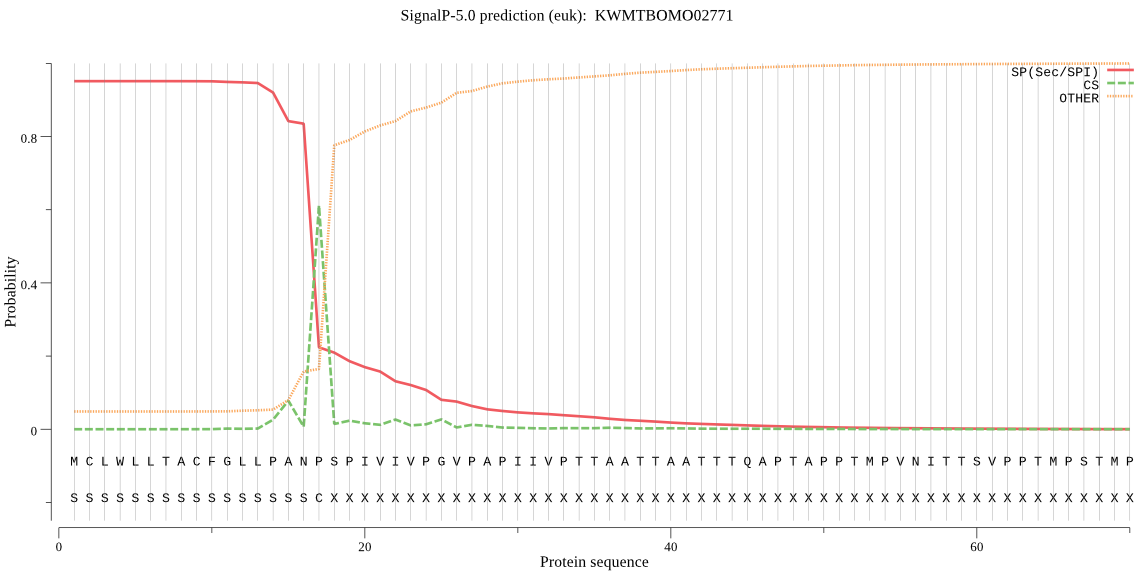
SignalP
Position: 1 - 17,
Likelihood: 0.950759
Length:
550
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.41333
Exp number, first 60 AAs:
0.91432
Total prob of N-in:
0.03825
outside
1 - 550
Population Genetic Test Statistics
Pi
207.437296
Theta
162.379062
Tajima's D
0.989082
CLR
0.343597
CSRT
0.651617419129044
Interpretation
Uncertain
