Gene
KWMTBOMO02767 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003709
Annotation
PREDICTED:_long-chain-fatty-acid--CoA_ligase_ACSBG2-like_isoform_X2_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.318 Mitochondrial Reliability : 1.998
Sequence
CDS
ATGGACGCGGTACAGGAACACAAAAACAATACGGAAAACTCAACGGTTACAGTTGAGAACGGTGCTACAGATACCCTTAAAACAAACAACATGCAAGTGGCGAATGGAGTCAGTCAACAAATATACTTAAACGGACCGGATCAAGTAGTGCCCGCAGAAAGTTACACAACAGCAATTCCTGGATGTCACGTCAAACTTCGCATCGCCCCCCGCGGCTTAGCAGCGGAGCCTCCTATATCAGTGCCTGGATTACTAAGTCGAACTACCGCCAGATATCCTAACGCTACAGCACTGGCCACAAAAAAGGCTGATGGGAAATGGCACAAAATCACCTACAAGCAATACCAGGATCGTGTCAGAATAATAGCAAAAGCTTTCTTGAAACTGGGTCTAGACAGGTACCATTCAGTCAGTATCCTCGGTTTCAACTCTGAACAATGGTTCATCGCCGATCTTGCAGCTATCCACGCTGGTGGGTATGCGGCGGGAATATATACAACCAACTCGGCCGACGCATGCTTCCACTGCCTCGAATCGTCGCGGGCTAACATCTGTGCCGTACAAGACAAGAAACAGCTGGACAAGATCCTTTCTGTGAAGCACAAGCTTCCCTTGTTGAAAGCCATCGTGCAATGGGAAGGACCTGTTGATACCTCCATACCTGGAATATATAGTTGGGACCAGCTACTAGAGATAGGAGCGAAGGAGCCAGACACGCAACTGAACGAAATTCTGAAAAGTATTGCCGTTAACGAATGCTGCACGTTGGTTTATACTTCAGGAACGGTGGGCCCGCCTAAGGCCGTGATGCTGTCTCACGATAACCTGACATGGGACGCGTTCGGCATCGGCGAGAGATGTCAGAATCTACAACCCACAAGAGACAGGCTCGTTTCATTCTTACCGCTCAGCCACGTTGCCGCTCAGGTAGTGGACATTTATACAACCTTGTCTAACGCTGTGACAGTGTACTTTGCACAACCAGACGCTCTGAAAGGCAGCCTCGTTGAAACCCTCAAAGAAGTCAGGCCAACAAGATTCTTGGGGGTCCCTCGGGTATGGGAAAAAATGTACGAGAAAATCATGGCCGTAGGTGCATCCAGTGGTCCACTTAAAAAGCAGATTGCTTTATGGGCCAAAGAGAAAGGGCTGCAATATCACCTCTCCAGGATTAACGGGTACGAGGGTTCGTCAGTTGGGTACAAGCTCGCTAAGTCTTTAGTGTTCAGCAAGATTCATGAGTCACTTGGCTTGGACAAGTGCAGCACTTTCGTAACGGCAGCTGCGCCACTGTCTCCCGACATCAAGAAATTCTTCCTTTCGTTGGATATCCCCCTTGTCGATGCTTTCGGAATGAGCGAGGCGGCCGGAGCCCACACCCTAAGTATTTATCCTAAGTTTTCTCTTGACTCTGCTGGCGAGATTTTGGAGGGAACCGAGACGAAATTCGGCGGCTCCATGAGTCCCAACGGTCCCGGTGAGATCATGATGAGAGGACGACACGTCTTTATGGGTTATTTGAACGACGCAGAAAAGACTAAAGGTGCTATCGACGATGATGGCTGGTTACTCTCCGGGGATGTTGGCAGAGTCGATTCGAACAATCTACTCTATATTACTGGTAGGATAAAGGAGCTGCTAATTACGGCCGGCGGCGAGAATGTGGCTCCAGTGCTGATCGAGCAAGCTATTCAAGCTGAACTCCTGCATGTAGGCTACGCTGTGCTCATTGGAGACCGACGAAAGTTCCTCTCTGTCTTGTTGACTCTGAAGACTAAGGTGAGTCCGGAGACGGGCGAGCCTTTAGATGAATTGGAAAGTGAAGCGAGGAAGTGGGTTGCTAGTTTGGGCAGTTCCGCAACTAAACTCAGTGAAATCGTTAACAGTAAAGACCCAGCGGTCCACAAAGCTATAGAGGCGGGTATCACACGCGCAAATAAGCATGCCATATCTAACGCTCAAAAAGTACAAAAGTTTGCGATCCTTCCCTCAGACTTCTCTGTGTATACCGGGGAATTGGGGCCAACATTAAAAATTAAGAGGAACGTCGTCTATGAGAAGTACAAAGACATAATTGAAGACTTTTACAAAGAGTGA
Protein
MDAVQEHKNNTENSTVTVENGATDTLKTNNMQVANGVSQQIYLNGPDQVVPAESYTTAIPGCHVKLRIAPRGLAAEPPISVPGLLSRTTARYPNATALATKKADGKWHKITYKQYQDRVRIIAKAFLKLGLDRYHSVSILGFNSEQWFIADLAAIHAGGYAAGIYTTNSADACFHCLESSRANICAVQDKKQLDKILSVKHKLPLLKAIVQWEGPVDTSIPGIYSWDQLLEIGAKEPDTQLNEILKSIAVNECCTLVYTSGTVGPPKAVMLSHDNLTWDAFGIGERCQNLQPTRDRLVSFLPLSHVAAQVVDIYTTLSNAVTVYFAQPDALKGSLVETLKEVRPTRFLGVPRVWEKMYEKIMAVGASSGPLKKQIALWAKEKGLQYHLSRINGYEGSSVGYKLAKSLVFSKIHESLGLDKCSTFVTAAAPLSPDIKKFFLSLDIPLVDAFGMSEAAGAHTLSIYPKFSLDSAGEILEGTETKFGGSMSPNGPGEIMMRGRHVFMGYLNDAEKTKGAIDDDGWLLSGDVGRVDSNNLLYITGRIKELLITAGGENVAPVLIEQAIQAELLHVGYAVLIGDRRKFLSVLLTLKTKVSPETGEPLDELESEARKWVASLGSSATKLSEIVNSKDPAVHKAIEAGITRANKHAISNAQKVQKFAILPSDFSVYTGELGPTLKIKRNVVYEKYKDIIEDFYKE
Summary
Uniprot
H9J2H2
A0A2H1WBR3
A0A212ETC2
A0A194QQ00
A0A194PN76
D6W7W1
+ More
A0A1B6E357 A0A2J7Q6C4 A0A2J7PZA5 A0A0L7RC40 K7J0T7 A0A232EWN7 A0A154NWY3 A0A067RJA3 A0A1L8DZG0 A0A1L8DYX0 A0A0C9QWM6 A0A1Q3FHG1 A0A1Q3FHD0 A0A2H8TZV7 E2A0J9 A0A1Q3FHI7 A0A0P4WGA2 A0A2A3EV16 A0A0J7K611 A0A088AJ72 J9JZ72 A0A182G222 A0A0T6AX18 A0A158NDN0 U5ETP1 A0A0P6J1B4 A0A026VZ11 A0A1S4JSS5 A0A195DBC6 A0A195DA07 A0A195BQA9 A0A182KMG2 A0A1S4GYP3 A0A2P2HVM7 A0A151XEH5 A0A182Q2C2 A0A182JF87 A0A2M3YZ03 W5LY77 A0A0P6BIV8 A0A182X5Z7 A0A0N8E9F9 A0A182LWE1 T1DPW4 A0A2M4BG99 A0A0P5EJR5 A0A0P5K6B5 A0A0P5KPF1 A0A0P4ZU62 A0A0N8ATF2 A0A0P5KDC8 A0A2M3YZ29 A0A0P6DJ83 A0A0P5NIA0 A0A0P5KA82 A0A2D0QW32 A0A0P5NHN4 E2C5D9 W5LY99 A0A1S3T5W5 B2GU97 A0A1S3T5V8 A0A034VJX4 A0A0P5HLP9 A0A0N8B0A2 A0A2M4A0Z1 A0A0K8UJF5 A0A1S3T5V9 A0A0N8DXD5 R0L2R8 Q7Q8J3 A0A0P5HZS1 U3I8S1 A0A195F3I3 A0A0A1XKY4 A0A182YQH4 A0A0P5Q6P9 F4WZ31 A0A0P5PF49 A0A3P8YES4 W8C978 A0A0P6DCA1 A0A195BP74 A0A0P5SJA2 A0A084VFF4 A0A3P8YDJ3 A0A182NTC4 A0A2M4CTV8 A0A2M4CTS3 A0A3P8YDI0 U3KHC6 W5LB37 A0A182WFK2 A0A060WEZ3
A0A1B6E357 A0A2J7Q6C4 A0A2J7PZA5 A0A0L7RC40 K7J0T7 A0A232EWN7 A0A154NWY3 A0A067RJA3 A0A1L8DZG0 A0A1L8DYX0 A0A0C9QWM6 A0A1Q3FHG1 A0A1Q3FHD0 A0A2H8TZV7 E2A0J9 A0A1Q3FHI7 A0A0P4WGA2 A0A2A3EV16 A0A0J7K611 A0A088AJ72 J9JZ72 A0A182G222 A0A0T6AX18 A0A158NDN0 U5ETP1 A0A0P6J1B4 A0A026VZ11 A0A1S4JSS5 A0A195DBC6 A0A195DA07 A0A195BQA9 A0A182KMG2 A0A1S4GYP3 A0A2P2HVM7 A0A151XEH5 A0A182Q2C2 A0A182JF87 A0A2M3YZ03 W5LY77 A0A0P6BIV8 A0A182X5Z7 A0A0N8E9F9 A0A182LWE1 T1DPW4 A0A2M4BG99 A0A0P5EJR5 A0A0P5K6B5 A0A0P5KPF1 A0A0P4ZU62 A0A0N8ATF2 A0A0P5KDC8 A0A2M3YZ29 A0A0P6DJ83 A0A0P5NIA0 A0A0P5KA82 A0A2D0QW32 A0A0P5NHN4 E2C5D9 W5LY99 A0A1S3T5W5 B2GU97 A0A1S3T5V8 A0A034VJX4 A0A0P5HLP9 A0A0N8B0A2 A0A2M4A0Z1 A0A0K8UJF5 A0A1S3T5V9 A0A0N8DXD5 R0L2R8 Q7Q8J3 A0A0P5HZS1 U3I8S1 A0A195F3I3 A0A0A1XKY4 A0A182YQH4 A0A0P5Q6P9 F4WZ31 A0A0P5PF49 A0A3P8YES4 W8C978 A0A0P6DCA1 A0A195BP74 A0A0P5SJA2 A0A084VFF4 A0A3P8YDJ3 A0A182NTC4 A0A2M4CTV8 A0A2M4CTS3 A0A3P8YDI0 U3KHC6 W5LB37 A0A182WFK2 A0A060WEZ3
Pubmed
EMBL
BABH01007945
BABH01007946
BABH01007947
BABH01007948
BABH01007949
BABH01007950
+ More
ODYU01007289 SOQ49934.1 AGBW02012631 OWR44701.1 KQ461181 KPJ07598.1 KQ459597 KPI94886.1 KQ971307 EFA11034.1 GEDC01006467 GEDC01004961 JAS30831.1 JAS32337.1 NEVH01017534 PNF24131.1 NEVH01020337 PNF21671.1 KQ414617 KOC68404.1 NNAY01001834 OXU22764.1 KQ434777 KZC04179.1 KK852474 KDR23088.1 GFDF01002412 JAV11672.1 GFDF01002413 JAV11671.1 GBYB01008139 JAG77906.1 GFDL01008057 JAV26988.1 GFDL01008113 JAV26932.1 GFXV01008050 MBW19855.1 GL435626 EFN72902.1 GFDL01008014 JAV27031.1 GDRN01090836 JAI60402.1 KZ288185 PBC34989.1 LBMM01013352 KMQ85684.1 ABLF02035823 JXUM01006539 JXUM01006540 JXUM01006541 LJIG01022614 KRT79624.1 ADTU01012724 ADTU01012725 GANO01001819 JAB58052.1 GDUN01000099 JAN95820.1 KK107578 EZA48686.1 KQ981082 KYN09719.1 KYN09718.1 KQ976424 KYM88318.1 AAAB01008944 IACF01000053 LAB65867.1 KQ982254 KYQ58786.1 AXCN02000715 GGFM01000742 MBW21493.1 AHAT01011275 AHAT01011276 GDIP01014366 JAM89349.1 GDIQ01048796 JAN45941.1 AXCM01001960 GAMD01002974 JAA98616.1 GGFJ01002906 MBW52047.1 GDIQ01268785 JAJ82939.1 GDIQ01188544 JAK63181.1 GDIQ01188546 GDIQ01188543 JAK63182.1 GDIP01209615 JAJ13787.1 GDIQ01244643 JAK07082.1 GDIQ01187046 JAK64679.1 GGFM01000773 MBW21524.1 GDIQ01245882 GDIQ01191250 GDIQ01189990 GDIQ01188545 GDIQ01099426 GDIQ01085799 LRGB01000568 JAN08938.1 KZS17817.1 GDIQ01268786 GDIQ01245884 GDIQ01245883 GDIQ01227629 GDIQ01189991 GDIQ01188547 GDIQ01187047 GDIQ01178345 GDIQ01178344 GDIQ01170448 GDIQ01169656 GDIQ01147778 GDIQ01147777 GDIQ01142017 GDIQ01103971 GDIQ01102675 GDIQ01087582 GDIQ01082589 GDIQ01065344 JAL09709.1 GDIQ01204645 GDIQ01187048 GDIQ01187045 JAK64680.1 GDIQ01142016 JAL09710.1 GL452770 EFN76872.1 BC166192 AAI66192.1 GAKP01016530 JAC42422.1 GDIQ01226333 JAK25392.1 GDIQ01225443 JAK26282.1 GGFK01001100 MBW34421.1 GDHF01032806 GDHF01032513 GDHF01025618 GDHF01014609 GDHF01013525 JAI19508.1 JAI19801.1 JAI26696.1 JAI37705.1 JAI38789.1 GDIQ01202841 GDIQ01082588 JAN12149.1 KB744699 EOA94577.1 EAA10184.3 GDIQ01222679 JAK29046.1 ADON01133919 KQ981820 KYN35140.1 GBXI01002742 JAD11550.1 GDIQ01125070 JAL26656.1 GL888465 EGI60534.1 GDIQ01129472 JAL22254.1 GAMC01006881 JAB99674.1 GDIQ01081038 JAN13699.1 KYM88317.1 GDIP01139077 JAL64637.1 ATLV01012386 KE524787 KFB36698.1 GGFL01004575 MBW68753.1 GGFL01004574 MBW68752.1 AGTO01021534 FR904519 CDQ65853.1
ODYU01007289 SOQ49934.1 AGBW02012631 OWR44701.1 KQ461181 KPJ07598.1 KQ459597 KPI94886.1 KQ971307 EFA11034.1 GEDC01006467 GEDC01004961 JAS30831.1 JAS32337.1 NEVH01017534 PNF24131.1 NEVH01020337 PNF21671.1 KQ414617 KOC68404.1 NNAY01001834 OXU22764.1 KQ434777 KZC04179.1 KK852474 KDR23088.1 GFDF01002412 JAV11672.1 GFDF01002413 JAV11671.1 GBYB01008139 JAG77906.1 GFDL01008057 JAV26988.1 GFDL01008113 JAV26932.1 GFXV01008050 MBW19855.1 GL435626 EFN72902.1 GFDL01008014 JAV27031.1 GDRN01090836 JAI60402.1 KZ288185 PBC34989.1 LBMM01013352 KMQ85684.1 ABLF02035823 JXUM01006539 JXUM01006540 JXUM01006541 LJIG01022614 KRT79624.1 ADTU01012724 ADTU01012725 GANO01001819 JAB58052.1 GDUN01000099 JAN95820.1 KK107578 EZA48686.1 KQ981082 KYN09719.1 KYN09718.1 KQ976424 KYM88318.1 AAAB01008944 IACF01000053 LAB65867.1 KQ982254 KYQ58786.1 AXCN02000715 GGFM01000742 MBW21493.1 AHAT01011275 AHAT01011276 GDIP01014366 JAM89349.1 GDIQ01048796 JAN45941.1 AXCM01001960 GAMD01002974 JAA98616.1 GGFJ01002906 MBW52047.1 GDIQ01268785 JAJ82939.1 GDIQ01188544 JAK63181.1 GDIQ01188546 GDIQ01188543 JAK63182.1 GDIP01209615 JAJ13787.1 GDIQ01244643 JAK07082.1 GDIQ01187046 JAK64679.1 GGFM01000773 MBW21524.1 GDIQ01245882 GDIQ01191250 GDIQ01189990 GDIQ01188545 GDIQ01099426 GDIQ01085799 LRGB01000568 JAN08938.1 KZS17817.1 GDIQ01268786 GDIQ01245884 GDIQ01245883 GDIQ01227629 GDIQ01189991 GDIQ01188547 GDIQ01187047 GDIQ01178345 GDIQ01178344 GDIQ01170448 GDIQ01169656 GDIQ01147778 GDIQ01147777 GDIQ01142017 GDIQ01103971 GDIQ01102675 GDIQ01087582 GDIQ01082589 GDIQ01065344 JAL09709.1 GDIQ01204645 GDIQ01187048 GDIQ01187045 JAK64680.1 GDIQ01142016 JAL09710.1 GL452770 EFN76872.1 BC166192 AAI66192.1 GAKP01016530 JAC42422.1 GDIQ01226333 JAK25392.1 GDIQ01225443 JAK26282.1 GGFK01001100 MBW34421.1 GDHF01032806 GDHF01032513 GDHF01025618 GDHF01014609 GDHF01013525 JAI19508.1 JAI19801.1 JAI26696.1 JAI37705.1 JAI38789.1 GDIQ01202841 GDIQ01082588 JAN12149.1 KB744699 EOA94577.1 EAA10184.3 GDIQ01222679 JAK29046.1 ADON01133919 KQ981820 KYN35140.1 GBXI01002742 JAD11550.1 GDIQ01125070 JAL26656.1 GL888465 EGI60534.1 GDIQ01129472 JAL22254.1 GAMC01006881 JAB99674.1 GDIQ01081038 JAN13699.1 KYM88317.1 GDIP01139077 JAL64637.1 ATLV01012386 KE524787 KFB36698.1 GGFL01004575 MBW68753.1 GGFL01004574 MBW68752.1 AGTO01021534 FR904519 CDQ65853.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000007266
UP000235965
+ More
UP000053825 UP000002358 UP000215335 UP000076502 UP000027135 UP000000311 UP000242457 UP000036403 UP000005203 UP000007819 UP000069940 UP000005205 UP000053097 UP000078492 UP000078540 UP000075882 UP000075809 UP000075886 UP000075880 UP000018468 UP000076407 UP000075883 UP000076858 UP000221080 UP000008237 UP000087266 UP000007062 UP000016666 UP000078541 UP000076408 UP000007755 UP000265140 UP000030765 UP000075884 UP000016665 UP000018467 UP000075920 UP000193380
UP000053825 UP000002358 UP000215335 UP000076502 UP000027135 UP000000311 UP000242457 UP000036403 UP000005203 UP000007819 UP000069940 UP000005205 UP000053097 UP000078492 UP000078540 UP000075882 UP000075809 UP000075886 UP000075880 UP000018468 UP000076407 UP000075883 UP000076858 UP000221080 UP000008237 UP000087266 UP000007062 UP000016666 UP000078541 UP000076408 UP000007755 UP000265140 UP000030765 UP000075884 UP000016665 UP000018467 UP000075920 UP000193380
PRIDE
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
ProteinModelPortal
H9J2H2
A0A2H1WBR3
A0A212ETC2
A0A194QQ00
A0A194PN76
D6W7W1
+ More
A0A1B6E357 A0A2J7Q6C4 A0A2J7PZA5 A0A0L7RC40 K7J0T7 A0A232EWN7 A0A154NWY3 A0A067RJA3 A0A1L8DZG0 A0A1L8DYX0 A0A0C9QWM6 A0A1Q3FHG1 A0A1Q3FHD0 A0A2H8TZV7 E2A0J9 A0A1Q3FHI7 A0A0P4WGA2 A0A2A3EV16 A0A0J7K611 A0A088AJ72 J9JZ72 A0A182G222 A0A0T6AX18 A0A158NDN0 U5ETP1 A0A0P6J1B4 A0A026VZ11 A0A1S4JSS5 A0A195DBC6 A0A195DA07 A0A195BQA9 A0A182KMG2 A0A1S4GYP3 A0A2P2HVM7 A0A151XEH5 A0A182Q2C2 A0A182JF87 A0A2M3YZ03 W5LY77 A0A0P6BIV8 A0A182X5Z7 A0A0N8E9F9 A0A182LWE1 T1DPW4 A0A2M4BG99 A0A0P5EJR5 A0A0P5K6B5 A0A0P5KPF1 A0A0P4ZU62 A0A0N8ATF2 A0A0P5KDC8 A0A2M3YZ29 A0A0P6DJ83 A0A0P5NIA0 A0A0P5KA82 A0A2D0QW32 A0A0P5NHN4 E2C5D9 W5LY99 A0A1S3T5W5 B2GU97 A0A1S3T5V8 A0A034VJX4 A0A0P5HLP9 A0A0N8B0A2 A0A2M4A0Z1 A0A0K8UJF5 A0A1S3T5V9 A0A0N8DXD5 R0L2R8 Q7Q8J3 A0A0P5HZS1 U3I8S1 A0A195F3I3 A0A0A1XKY4 A0A182YQH4 A0A0P5Q6P9 F4WZ31 A0A0P5PF49 A0A3P8YES4 W8C978 A0A0P6DCA1 A0A195BP74 A0A0P5SJA2 A0A084VFF4 A0A3P8YDJ3 A0A182NTC4 A0A2M4CTV8 A0A2M4CTS3 A0A3P8YDI0 U3KHC6 W5LB37 A0A182WFK2 A0A060WEZ3
A0A1B6E357 A0A2J7Q6C4 A0A2J7PZA5 A0A0L7RC40 K7J0T7 A0A232EWN7 A0A154NWY3 A0A067RJA3 A0A1L8DZG0 A0A1L8DYX0 A0A0C9QWM6 A0A1Q3FHG1 A0A1Q3FHD0 A0A2H8TZV7 E2A0J9 A0A1Q3FHI7 A0A0P4WGA2 A0A2A3EV16 A0A0J7K611 A0A088AJ72 J9JZ72 A0A182G222 A0A0T6AX18 A0A158NDN0 U5ETP1 A0A0P6J1B4 A0A026VZ11 A0A1S4JSS5 A0A195DBC6 A0A195DA07 A0A195BQA9 A0A182KMG2 A0A1S4GYP3 A0A2P2HVM7 A0A151XEH5 A0A182Q2C2 A0A182JF87 A0A2M3YZ03 W5LY77 A0A0P6BIV8 A0A182X5Z7 A0A0N8E9F9 A0A182LWE1 T1DPW4 A0A2M4BG99 A0A0P5EJR5 A0A0P5K6B5 A0A0P5KPF1 A0A0P4ZU62 A0A0N8ATF2 A0A0P5KDC8 A0A2M3YZ29 A0A0P6DJ83 A0A0P5NIA0 A0A0P5KA82 A0A2D0QW32 A0A0P5NHN4 E2C5D9 W5LY99 A0A1S3T5W5 B2GU97 A0A1S3T5V8 A0A034VJX4 A0A0P5HLP9 A0A0N8B0A2 A0A2M4A0Z1 A0A0K8UJF5 A0A1S3T5V9 A0A0N8DXD5 R0L2R8 Q7Q8J3 A0A0P5HZS1 U3I8S1 A0A195F3I3 A0A0A1XKY4 A0A182YQH4 A0A0P5Q6P9 F4WZ31 A0A0P5PF49 A0A3P8YES4 W8C978 A0A0P6DCA1 A0A195BP74 A0A0P5SJA2 A0A084VFF4 A0A3P8YDJ3 A0A182NTC4 A0A2M4CTV8 A0A2M4CTS3 A0A3P8YDI0 U3KHC6 W5LB37 A0A182WFK2 A0A060WEZ3
PDB
5MSW
E-value=8.25447e-20,
Score=241
Ontologies
PATHWAY
GO
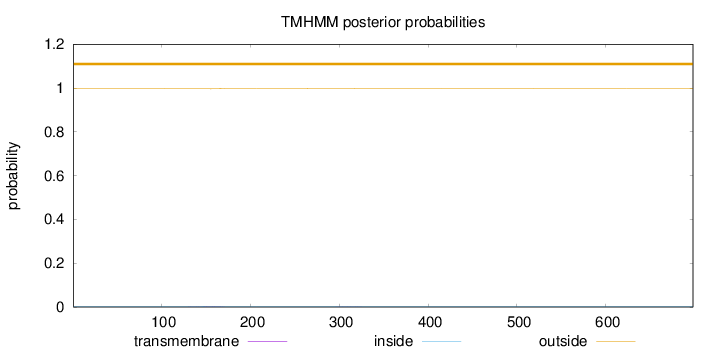
Topology
Length:
698
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0661600000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00372
outside
1 - 698
Population Genetic Test Statistics
Pi
207.935277
Theta
182.274497
Tajima's D
0.613889
CLR
0.82161
CSRT
0.544072796360182
Interpretation
Uncertain
