Gene
KWMTBOMO02766
Pre Gene Modal
BGIBMGA003488
Annotation
PREDICTED:_patj_homolog_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.151
Sequence
CDS
ATGCATTTGGGCGTGAACGTCTCAAATGCATTACAGCAATTGGAAAGTGTGAAAGCTGCTGTGGACCAGAGCAATGATCCCAAGCTTAAAGCTGCAACCAGTGATGATTTGAACATGCTAATAAGTTTATTAGAGAGTCCCATATTACGAAGTATTACTACACTTCATGACTCTGTGGGCATGCTTGCAACTCAGGTTGCACATCACCCATCAATTTTACCTGGAGATTTTGATATTACTCCTGCTGGTGACTTGGCATTACAAACAAGAAATTTGTATGGAAATCAAGAGGGTGAAGAGGAGCAAAGGGTGCCACAGGTCTCACCTCCACATAGTATGGAGTTTGGATCTGATAATGAACATATTCTTGGACTTGATAGTGGCTCTGTTGACTCTAACATGTCTCCTAAACAAAGTCGTGGCTTATTAGATTTGTCTCGTAATGATGATTCTTTAGCATCTACCAATCACAACGTAGTAGCAGGGGACTGGGCACAAGTTGAAATTATAAATCTTGTAAATGATGGAACTGGCCTTGGATTTGGTATTATAGGTGCTAGAACAAGTGGTGTCATTGTAAAGACGATACTAGCAGGTGGTGTAGCTGATCGCGATGGCCGACTAAAATCTGGAGATCACATTCTACAGATTGGAGATGTAAGCCTTATGGAGATGGGTTCCGAGCAAGTAGCTGGTGTGCTACGTGCGTCCGGTAGTCGTGTGAGGCTCGTCGTGGCGCGAGCAGTGGATCCGGCGGCCTTGCATGCGTCACCCGCTCCCGTCGTGCCTGCCAGGCTTTTGTCAGACCCCGAATCGCTGGATCGCTATCTCATGGAAGCTGGTTTTGAGCAAGTGTTCCATACGCAGCCTGTGCCTGTCAGCTCCACTTCGACGAGATTTGTATTTGACAATCCCATAACACCCCCTCCAGATTCAGATGCTGAATCACCAGAAGTTGATAGATTCACAGTTCAACTGAAGAAAGACGAGAACGGCTTAGGGATTACTATTGCAGGTTATGTTTGCGAAAAGGAGGAGCTGTCAGGTATATTTGTGAAGTCGGTGACTCCGGGGAGCGCTGCAGCGTTGAGCGGAATGGTGAGGGTCAATGACCGCATCGTTGCCGTCGATGGCGTCTCCCTCGCAGGGAAATCCAACCAGCGCGCTGTGGACGCCCTGAAACAGAGTCGCAACGTTGTCACGCTAGAACTAGAAAGATACCTGCGTGGTCCGAAATATGAGCAACTGCAGCAAGCGATAGCGGCGGGCGAGGCGGCCGCCCCCCCTCCCCCGCCCAACCCGTTCCTGCACATGCACCGTCCCGACGCACCTTCTCCTGAAATAGAAATACAGCTCTCCAACCTCCAAATAACGGATAACGGCACAGATGACGCTGTGCCTCTAACTCGCAGCAGTCCTCCGGCGACTGCACTGCGTCCGGCACCAGCGTTCGAGATGCCACGGACTCCTGAACAAAAAGAAGCGATAAGAAGGAAATGGCAGGCTATATTAGGCGAGGACGTGGAGATAGTGGTGGGCGTGGTGGTGCGCGGTGGGGGTGGTCTGGGCATTTCGCTAGAAGGCACCGTGGACGTGGAGGGGGGGCGCGAGGTGAGGCCGCATCACTACGTGCGCTCCGTGCTGCCCGAGGGGCCCGTGGGCCGCGCCGCCGTGCATCGCCCCGGCGACGAGCTGCTCGAAGTGAACGGTCATCGGCTCCTGGGCATGAACCACTTAGAAGTGGTGTCAATTCTGAAGGAATTATCGAGTGAAGTCTGCATGGTGTGTGCGAGACCTCGACCTGCGCCCCCTCTCGACCTAGCGCCGCCTGCCACAACATTAGTGAAGGCGAAATCGGACGGTAGCCTGGCGGGTGCGGGTGCTGAGGAAGGCGGGTCGCTGTCCGCCGCTGGGAAAGTTCGTTCCCGGAGTCTCGAACCACTAACCGGACTTGCTATGTGGTCTTCTGAACCCCAGATAATAGAGCTAGTGAAAGGCGAGCGTGGTCTGGGGTTCTCGATCCTGGACTACCAGGACCCGCTGCGGCCCGCGCACACGCTCGTCGTCATCCGCTCGCTGGTGCCGGGCGGCGTGGCGCAGCAGGACGGCCGCCTCATACCCGGGGACAGGCTGCTCTTCGTTAACGATCAGAATCTTGAAAATGCGAGTCTAGAGCAAGCGGTGGCGGCGCTGAAGGGCGCGCCCCGAGGAGTGGTCCGCATCGGGGTGGCCAAGCCTCTACCGCTGGCCGACGCGCCGCCGCCGCTGCCCACCACGCCGCCCCCCAACACCTTGAGCTATAAAATATTTACTATGAACTAG
Protein
MHLGVNVSNALQQLESVKAAVDQSNDPKLKAATSDDLNMLISLLESPILRSITTLHDSVGMLATQVAHHPSILPGDFDITPAGDLALQTRNLYGNQEGEEEQRVPQVSPPHSMEFGSDNEHILGLDSGSVDSNMSPKQSRGLLDLSRNDDSLASTNHNVVAGDWAQVEIINLVNDGTGLGFGIIGARTSGVIVKTILAGGVADRDGRLKSGDHILQIGDVSLMEMGSEQVAGVLRASGSRVRLVVARAVDPAALHASPAPVVPARLLSDPESLDRYLMEAGFEQVFHTQPVPVSSTSTRFVFDNPITPPPDSDAESPEVDRFTVQLKKDENGLGITIAGYVCEKEELSGIFVKSVTPGSAAALSGMVRVNDRIVAVDGVSLAGKSNQRAVDALKQSRNVVTLELERYLRGPKYEQLQQAIAAGEAAAPPPPPNPFLHMHRPDAPSPEIEIQLSNLQITDNGTDDAVPLTRSSPPATALRPAPAFEMPRTPEQKEAIRRKWQAILGEDVEIVVGVVVRGGGGLGISLEGTVDVEGGREVRPHHYVRSVLPEGPVGRAAVHRPGDELLEVNGHRLLGMNHLEVVSILKELSSEVCMVCARPRPAPPLDLAPPATTLVKAKSDGSLAGAGAEEGGSLSAAGKVRSRSLEPLTGLAMWSSEPQIIELVKGERGLGFSILDYQDPLRPAHTLVVIRSLVPGGVAQQDGRLIPGDRLLFVNDQNLENASLEQAVAALKGAPRGVVRIGVAKPLPLADAPPPLPTTPPPNTLSYKIFTMN
Summary
Uniprot
A0A2A4K1J6
A0A2H1WVM7
H9J1V1
A0A212ET84
A0A194PUS4
A0A194QVL6
+ More
A0A1Y1MNC1 D6W6M5 A0A1J1I5Y6 A0A1W4WZU9 E0VGE8 A0A146LAK9 A0A146KZ93 A0A1B6ELN7 A0A2J7R5D2 A0A2J7R5D3 A0A0P4VIU4 A0A023F3W4 A0A195BWA7 A0A0M8ZVG3 A0A151JQU8 A0A1B6GQS6 A0A154PNZ1 A0A3L8DGH0 A0A026X228 A0A232FA68 E2B674 A0A0A9YHT4 A0A2J7R5D1 A0A0L7R1E0 A0A2J7R5D6 F4WWZ8 A0A151WMY2 A0A224X4S0 A0A087ZV81 T1INI3 A0A151IBX1 A0A1Y1M2Z1 A0A2W1BUE1 A0A2A4JGR2 A0A0T6ASQ8 A0A2H1V279 A0A195F8V7 A0A0K2SZ76 E9INJ5 A0A2H5BFC6 H3AIS3
A0A1Y1MNC1 D6W6M5 A0A1J1I5Y6 A0A1W4WZU9 E0VGE8 A0A146LAK9 A0A146KZ93 A0A1B6ELN7 A0A2J7R5D2 A0A2J7R5D3 A0A0P4VIU4 A0A023F3W4 A0A195BWA7 A0A0M8ZVG3 A0A151JQU8 A0A1B6GQS6 A0A154PNZ1 A0A3L8DGH0 A0A026X228 A0A232FA68 E2B674 A0A0A9YHT4 A0A2J7R5D1 A0A0L7R1E0 A0A2J7R5D6 F4WWZ8 A0A151WMY2 A0A224X4S0 A0A087ZV81 T1INI3 A0A151IBX1 A0A1Y1M2Z1 A0A2W1BUE1 A0A2A4JGR2 A0A0T6ASQ8 A0A2H1V279 A0A195F8V7 A0A0K2SZ76 E9INJ5 A0A2H5BFC6 H3AIS3
Pubmed
EMBL
NWSH01000300
PCG77633.1
ODYU01011401
SOQ57125.1
BABH01007952
BABH01007953
+ More
AGBW02012631 OWR44702.1 KQ459597 KPI94885.1 KQ461181 KPJ07596.1 GEZM01027922 JAV86578.1 KQ971307 EFA10862.2 CVRI01000040 CRK94996.1 DS235140 EEB12454.1 GDHC01014477 JAQ04152.1 GDHC01018599 JAQ00030.1 GECZ01030919 JAS38850.1 NEVH01006994 PNF36041.1 PNF36042.1 GDKW01001773 JAI54822.1 GBBI01002582 JAC16130.1 KQ976396 KYM92914.1 KQ435830 KOX71767.1 KQ978619 KYN29768.1 GECZ01004998 JAS64771.1 KQ435007 KZC13605.1 QOIP01000008 RLU19272.1 KK107034 EZA62056.1 NNAY01000618 OXU27370.1 GL445930 EFN88767.1 GBHO01012413 GBRD01008386 JAG31191.1 JAG57435.1 PNF36039.1 KQ414668 KOC64659.1 PNF36040.1 GL888417 EGI61241.1 KQ982934 KYQ49168.1 GFTR01008919 JAW07507.1 JH431176 KQ978076 KYM97225.1 GEZM01041885 GEZM01041880 JAV80119.1 KZ149945 PZC76817.1 NWSH01001542 PCG70908.1 LJIG01022905 KRT78129.1 ODYU01000125 SOQ34374.1 KQ981727 KYN37040.1 HACA01001326 CDW18687.1 GL764306 EFZ17853.1 MG197681 AUG84431.1 AFYH01165142 AFYH01165143 AFYH01165144 AFYH01165145 AFYH01165146 AFYH01165147 AFYH01165148 AFYH01165149 AFYH01165150 AFYH01165151
AGBW02012631 OWR44702.1 KQ459597 KPI94885.1 KQ461181 KPJ07596.1 GEZM01027922 JAV86578.1 KQ971307 EFA10862.2 CVRI01000040 CRK94996.1 DS235140 EEB12454.1 GDHC01014477 JAQ04152.1 GDHC01018599 JAQ00030.1 GECZ01030919 JAS38850.1 NEVH01006994 PNF36041.1 PNF36042.1 GDKW01001773 JAI54822.1 GBBI01002582 JAC16130.1 KQ976396 KYM92914.1 KQ435830 KOX71767.1 KQ978619 KYN29768.1 GECZ01004998 JAS64771.1 KQ435007 KZC13605.1 QOIP01000008 RLU19272.1 KK107034 EZA62056.1 NNAY01000618 OXU27370.1 GL445930 EFN88767.1 GBHO01012413 GBRD01008386 JAG31191.1 JAG57435.1 PNF36039.1 KQ414668 KOC64659.1 PNF36040.1 GL888417 EGI61241.1 KQ982934 KYQ49168.1 GFTR01008919 JAW07507.1 JH431176 KQ978076 KYM97225.1 GEZM01041885 GEZM01041880 JAV80119.1 KZ149945 PZC76817.1 NWSH01001542 PCG70908.1 LJIG01022905 KRT78129.1 ODYU01000125 SOQ34374.1 KQ981727 KYN37040.1 HACA01001326 CDW18687.1 GL764306 EFZ17853.1 MG197681 AUG84431.1 AFYH01165142 AFYH01165143 AFYH01165144 AFYH01165145 AFYH01165146 AFYH01165147 AFYH01165148 AFYH01165149 AFYH01165150 AFYH01165151
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000053240
UP000007266
+ More
UP000183832 UP000192223 UP000009046 UP000235965 UP000078540 UP000053105 UP000078492 UP000076502 UP000279307 UP000053097 UP000215335 UP000008237 UP000053825 UP000007755 UP000075809 UP000005203 UP000078542 UP000078541 UP000008672
UP000183832 UP000192223 UP000009046 UP000235965 UP000078540 UP000053105 UP000078492 UP000076502 UP000279307 UP000053097 UP000215335 UP000008237 UP000053825 UP000007755 UP000075809 UP000005203 UP000078542 UP000078541 UP000008672
PRIDE
Interpro
ProteinModelPortal
A0A2A4K1J6
A0A2H1WVM7
H9J1V1
A0A212ET84
A0A194PUS4
A0A194QVL6
+ More
A0A1Y1MNC1 D6W6M5 A0A1J1I5Y6 A0A1W4WZU9 E0VGE8 A0A146LAK9 A0A146KZ93 A0A1B6ELN7 A0A2J7R5D2 A0A2J7R5D3 A0A0P4VIU4 A0A023F3W4 A0A195BWA7 A0A0M8ZVG3 A0A151JQU8 A0A1B6GQS6 A0A154PNZ1 A0A3L8DGH0 A0A026X228 A0A232FA68 E2B674 A0A0A9YHT4 A0A2J7R5D1 A0A0L7R1E0 A0A2J7R5D6 F4WWZ8 A0A151WMY2 A0A224X4S0 A0A087ZV81 T1INI3 A0A151IBX1 A0A1Y1M2Z1 A0A2W1BUE1 A0A2A4JGR2 A0A0T6ASQ8 A0A2H1V279 A0A195F8V7 A0A0K2SZ76 E9INJ5 A0A2H5BFC6 H3AIS3
A0A1Y1MNC1 D6W6M5 A0A1J1I5Y6 A0A1W4WZU9 E0VGE8 A0A146LAK9 A0A146KZ93 A0A1B6ELN7 A0A2J7R5D2 A0A2J7R5D3 A0A0P4VIU4 A0A023F3W4 A0A195BWA7 A0A0M8ZVG3 A0A151JQU8 A0A1B6GQS6 A0A154PNZ1 A0A3L8DGH0 A0A026X228 A0A232FA68 E2B674 A0A0A9YHT4 A0A2J7R5D1 A0A0L7R1E0 A0A2J7R5D6 F4WWZ8 A0A151WMY2 A0A224X4S0 A0A087ZV81 T1INI3 A0A151IBX1 A0A1Y1M2Z1 A0A2W1BUE1 A0A2A4JGR2 A0A0T6ASQ8 A0A2H1V279 A0A195F8V7 A0A0K2SZ76 E9INJ5 A0A2H5BFC6 H3AIS3
PDB
2DLU
E-value=6.9451e-24,
Score=277
Ontologies
GO
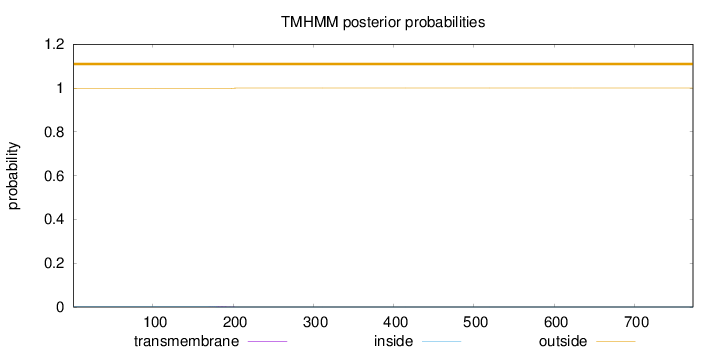
Topology
Length:
773
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0323100000000002
Exp number, first 60 AAs:
0.00075
Total prob of N-in:
0.00147
outside
1 - 773
Population Genetic Test Statistics
Pi
251.254328
Theta
180.208689
Tajima's D
1.31475
CLR
0.045423
CSRT
0.742012899355032
Interpretation
Uncertain
