Gene
KWMTBOMO02753
Pre Gene Modal
BGIBMGA003715
Annotation
nicotinic_acetylcholine_receptor_subunit_alpha_7_isoform_2_precursor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.575
Sequence
CDS
ATGGGCGGGCGGGCGCGCTGCTCGCTACTGGCGGCGCCCGCAGGCCTTCTGCTACTACTGCTGGGCCTGCTGTGGCCGAGGGGGGTGTGCGGCGGTCACCACGAGAAGCGGTTGTTGCACCATCTGCTGGACCACTACAACGTGCTGGAGCGCCCTGTGGTCAACGAGAGCGACCCGCTGCAGCTGTCCTTCGGCCTCACGCTCATGCAGATCATTGACGTGGATGAAAAGAACCAGCTTTTGATAACAAACATTTGGCTCAAATTAGAATGGAATGATATGAATTTGAGGTGGAACACTTCAGATTTCGGCGGAGTTAAAGATTTAAGAGTACCACCTCACAGACTATGGAAACCAGATGTTCTTATGTACAACAGTGCGGACGAAGGCTTCGACAGCACGTACCCAACCAATGTTGTGGTGCGGAACAACGGCTCGTGCCTTTACGTCCCACCCGGCATATTCAAGAGCACATGCAAAATAGACATCACGTGGTTTCCATTCGATGACCAACGCTGCGAAATGAAGTTTGGCAGTTGGACATATGACGGCTATCAATTGGATCTTCAATTACAAGATGAAGCGGGTGGGGATATAAGCAGTTTTGTCACGAATGGCGAGTGGGAGTTAATCGGAGTACCAGGCAAGCGGAACGAAATCTATTACAATTGCTGCCCAGAACCGTATATAGACATAACATTCGCGGTCGTTATCCGTCGAAAAACGTTATACTATTTCTTCAATCTCATAGTACCGTGTGTCTTAATCGCATCAATGGCGCTACTCGGGTTCACACTGCCACCGGATTCTGGAGAAAAGCTATCTTTAGGTGTCACAATTCTATTGTCGTTGACGGTGTTTCTGAACATGGTGGCCGAAACGATGCCCGCAACATCAGATGCTGTGCCTCTGCTCGGTACTTACTTCAACTGCATCATGTTCATGGTCGCCTCTTCCGTGGTCTCCACTATACTCATCCTGAATTATCACCACAGACATGCAGACACGCATGAAATGAGTGACTGGATCCGGTGTGTGTTTCTGTACTGGTTGCCTTGGATCCTGCGTATGTCAAGGCCGGGTTCGGCGACGACGCCGCCGCCGGCTCGAGCGCCCCCCCCTCCCGACCTGGAGCTACGTGAGCGCTCCTCCAAGTCACTCCTCGCTAACGTGCTCGACATCGACGACGACTTCCGGCACGCGCAATCGCAGCAGCCGCCCTGCTGCCGCTACTACAGGGGGGGCGAAGAAAATGGAGCAGGCTTAGCGGCGCACAGCTGTTTCGGTGTCGACTACGAGCTGTCGCTCATATTGAAGGAAATCCGAGTCATCACAGATCAGATGCGCAAAGATGACGAAGATGCGGACATATCGCGCGACTGGAAATTTGCCGCCATGGTCGTGGACAGACTGTGCCTTATTATCTTTACCCTGTTCACAATCATCGCCACGCTAGCCGTGCTGCTGTCCGCGCCGCACATCATGGTGTCGTAG
Protein
MGGRARCSLLAAPAGLLLLLLGLLWPRGVCGGHHEKRLLHHLLDHYNVLERPVVNESDPLQLSFGLTLMQIIDVDEKNQLLITNIWLKLEWNDMNLRWNTSDFGGVKDLRVPPHRLWKPDVLMYNSADEGFDSTYPTNVVVRNNGSCLYVPPGIFKSTCKIDITWFPFDDQRCEMKFGSWTYDGYQLDLQLQDEAGGDISSFVTNGEWELIGVPGKRNEIYYNCCPEPYIDITFAVVIRRKTLYYFFNLIVPCVLIASMALLGFTLPPDSGEKLSLGVTILLSLTVFLNMVAETMPATSDAVPLLGTYFNCIMFMVASSVVSTILILNYHHRHADTHEMSDWIRCVFLYWLPWILRMSRPGSATTPPPARAPPPPDLELRERSSKSLLANVLDIDDDFRHAQSQQPPCCRYYRGGEENGAGLAAHSCFGVDYELSLILKEIRVITDQMRKDDEDADISRDWKFAAMVVDRLCLIIFTLFTIIATLAVLLSAPHIMVS
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A8CGM5
A8HTC8
A0A0H4TRG2
A8HTC5
Q9XZI3
A0A0H4TQC6
+ More
A0A0B5GNX1 A0A194RGP8 A0A2H1W7E5 A0A212FIW5 A0A2A4K153 A0A2A4K033 A0A346IHV2 B4JD77 A0A1S4G7N3 A0A084VFG0 A0A1S4GZF8 Q29KA5 Q7PHY7 Q8T7V5 B3MMS1 A0A084VN01 A0A0J9R2A0 Q7KT97 B4M8Q7 B4P0I6 B3NA88 A0A1W4UXW1 Q7QI13 A0A1J1IWW5 A8DIU0 A8HTF2 B4MVM6 A9XFY4 A0A0L7LQI0 B4Q5H4 B4KEK2 B4HXE7 A0A0L0BUB3 A0A240SWJ8 E1JJR2 A0A0R1EB74 Q86MN7 B3NVM9 Q9VWI9 A0A3B0KMX8 B4PZJ0 B4MGC3 A4GXC7 A0A1W4VG42 B4L7L9 W8B2K3 A0A0C5I2P3 A0A348BSW2 B8X4G9 B4JJS4 A0A1W4X2L4 A0A348BSW4 A0A1Y1MEU7 A0A023GVG5 A0A348BSW3 A0A1D8GS61 G9CU47 K4JEQ6 A0A0R3P102 A0A3B0KA18 A0A0P5EP72 A0A2S2PZX2 A0A162CYS9 A0A0U2GQ98 A0A1I8PTD7 J9JT17 E9GTK4 A0A2J7PVE3 A0A1I8PTI3 W6EL40 A0A1I8PNY4 A0A182KMG8 A0A2A4JGH6 A0A2W1BAC7 A0A0H4TMF2 A0A158NJP1 A0A0Q9X676 K4JF52 A0A0H4U7U0 A8CGL6 A0A1I8NYF7 A0A0P8ZHV2 G9CU48 A0A0Q9W8H3 A5H032
A0A0B5GNX1 A0A194RGP8 A0A2H1W7E5 A0A212FIW5 A0A2A4K153 A0A2A4K033 A0A346IHV2 B4JD77 A0A1S4G7N3 A0A084VFG0 A0A1S4GZF8 Q29KA5 Q7PHY7 Q8T7V5 B3MMS1 A0A084VN01 A0A0J9R2A0 Q7KT97 B4M8Q7 B4P0I6 B3NA88 A0A1W4UXW1 Q7QI13 A0A1J1IWW5 A8DIU0 A8HTF2 B4MVM6 A9XFY4 A0A0L7LQI0 B4Q5H4 B4KEK2 B4HXE7 A0A0L0BUB3 A0A240SWJ8 E1JJR2 A0A0R1EB74 Q86MN7 B3NVM9 Q9VWI9 A0A3B0KMX8 B4PZJ0 B4MGC3 A4GXC7 A0A1W4VG42 B4L7L9 W8B2K3 A0A0C5I2P3 A0A348BSW2 B8X4G9 B4JJS4 A0A1W4X2L4 A0A348BSW4 A0A1Y1MEU7 A0A023GVG5 A0A348BSW3 A0A1D8GS61 G9CU47 K4JEQ6 A0A0R3P102 A0A3B0KA18 A0A0P5EP72 A0A2S2PZX2 A0A162CYS9 A0A0U2GQ98 A0A1I8PTD7 J9JT17 E9GTK4 A0A2J7PVE3 A0A1I8PTI3 W6EL40 A0A1I8PNY4 A0A182KMG8 A0A2A4JGH6 A0A2W1BAC7 A0A0H4TMF2 A0A158NJP1 A0A0Q9X676 K4JF52 A0A0H4U7U0 A8CGL6 A0A1I8NYF7 A0A0P8ZHV2 G9CU48 A0A0Q9W8H3 A5H032
Pubmed
17868469
19121390
26354079
22118469
17994087
24438588
+ More
12364791 15632085 14747013 15676276 17210077 11973307 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 17880682 18362917 19320762 19820115 18092998 26227816 26108605 16494528 24495485 28004739 23185243 21292972 25701599 20966253 28756777 21347285 17439546
12364791 15632085 14747013 15676276 17210077 11973307 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 17880682 18362917 19320762 19820115 18092998 26227816 26108605 16494528 24495485 28004739 23185243 21292972 25701599 20966253 28756777 21347285 17439546
EMBL
BABH01007977
BABH01007978
EU082084
ABV45520.2
EU143798
ABV72689.1
+ More
KP711056 AKQ12752.1 EU143797 ABV72688.1 AF143846 AAD32697.1 KP711049 RSAL01000017 AKQ12745.1 RVE52946.1 KM884875 AJF23047.1 KQ460207 KPJ16762.1 ODYU01006685 SOQ48752.1 AGBW02008321 OWR53672.1 NWSH01000300 PCG77624.1 PCG77625.1 MH138166 AXP17107.1 CH916368 EDW03250.1 ATLV01012387 ATLV01012388 KE524787 KFB36704.1 AAAB01008944 CH379061 EAL33270.2 AY705399 AAU12508.1 EAA44351.2 AF272778 AAM13390.1 CH902620 EDV31962.1 ATLV01014657 ATLV01014658 KE524978 KFB39345.1 CM002910 KMY90213.1 AE014134 AAS64702.2 AAS64703.1 AFH03701.1 CH940654 EDW57583.2 CM000157 EDW88980.1 CH954177 EDV58590.1 KQS70451.1 KQS70453.1 AY705402 AAAB01008811 AAU12511.1 EAA04911.3 CVRI01000063 CRL04701.1 EF526089 EU937796 KQ971307 ABS86911.1 ACP31301.1 EFA12057.1 EU143806 ABV72697.1 CH963857 EDW75746.2 EF203213 ABY40460.1 JTDY01000311 KOB77753.1 CM000361 EDX05018.1 CH933807 EDW11881.2 CH480818 EDW51727.1 JRES01001455 KNC22799.1 AE014298 ACZ95329.1 CM000162 KRK06589.1 AJ554210 CAD86936.1 CH954180 EDV46694.2 AAF48950.4 AAS65409.1 AHN59906.1 OUUW01000011 SPP86491.1 EDX02146.2 CH940672 EDW57446.2 EF458626 ABO26063.1 CH933814 EDW05732.2 GAMC01019089 GAMC01019088 JAB87466.1 KP090397 AJP36493.1 LC389186 BBE49565.1 FJ167396 ACL14949.1 CH916370 EDV99826.1 LC389188 BBE49567.1 GEZM01036763 JAV82396.1 JQ973830 AFN73052.1 LC389187 BBE49566.1 KU557781 AOT81843.1 JF731242 AEA40428.1 JX466891 AFU82562.1 KRT04867.1 KRT04868.1 KRT04869.1 OUUW01000006 SPP82496.1 GDIQ01269329 JAJ82395.1 GGMS01001872 MBY71075.1 LRGB01000024 KZS21319.1 KT328074 ALB27752.1 ABLF02027371 ABLF02027373 GL732564 EFX77198.1 NEVH01020963 PNF20307.1 KF873587 AHJ11216.1 NWSH01001615 PCG70684.1 KZ150420 PZC70934.1 KP711055 AKQ12751.1 ADTU01018111 ADTU01018112 ADTU01018113 ADTU01018114 ADTU01018115 ADTU01018116 ADTU01018117 ADTU01018118 ADTU01018119 ADTU01018120 KRG03750.1 JX466888 AFU82559.1 KP711048 AKQ12744.1 EU082082 ABV45518.1 CH902624 KPU74285.1 JF731243 KP725465 AEA40429.1 AKV94622.1 CH940649 KRF81069.1 DQ498130 ABJ09669.1
KP711056 AKQ12752.1 EU143797 ABV72688.1 AF143846 AAD32697.1 KP711049 RSAL01000017 AKQ12745.1 RVE52946.1 KM884875 AJF23047.1 KQ460207 KPJ16762.1 ODYU01006685 SOQ48752.1 AGBW02008321 OWR53672.1 NWSH01000300 PCG77624.1 PCG77625.1 MH138166 AXP17107.1 CH916368 EDW03250.1 ATLV01012387 ATLV01012388 KE524787 KFB36704.1 AAAB01008944 CH379061 EAL33270.2 AY705399 AAU12508.1 EAA44351.2 AF272778 AAM13390.1 CH902620 EDV31962.1 ATLV01014657 ATLV01014658 KE524978 KFB39345.1 CM002910 KMY90213.1 AE014134 AAS64702.2 AAS64703.1 AFH03701.1 CH940654 EDW57583.2 CM000157 EDW88980.1 CH954177 EDV58590.1 KQS70451.1 KQS70453.1 AY705402 AAAB01008811 AAU12511.1 EAA04911.3 CVRI01000063 CRL04701.1 EF526089 EU937796 KQ971307 ABS86911.1 ACP31301.1 EFA12057.1 EU143806 ABV72697.1 CH963857 EDW75746.2 EF203213 ABY40460.1 JTDY01000311 KOB77753.1 CM000361 EDX05018.1 CH933807 EDW11881.2 CH480818 EDW51727.1 JRES01001455 KNC22799.1 AE014298 ACZ95329.1 CM000162 KRK06589.1 AJ554210 CAD86936.1 CH954180 EDV46694.2 AAF48950.4 AAS65409.1 AHN59906.1 OUUW01000011 SPP86491.1 EDX02146.2 CH940672 EDW57446.2 EF458626 ABO26063.1 CH933814 EDW05732.2 GAMC01019089 GAMC01019088 JAB87466.1 KP090397 AJP36493.1 LC389186 BBE49565.1 FJ167396 ACL14949.1 CH916370 EDV99826.1 LC389188 BBE49567.1 GEZM01036763 JAV82396.1 JQ973830 AFN73052.1 LC389187 BBE49566.1 KU557781 AOT81843.1 JF731242 AEA40428.1 JX466891 AFU82562.1 KRT04867.1 KRT04868.1 KRT04869.1 OUUW01000006 SPP82496.1 GDIQ01269329 JAJ82395.1 GGMS01001872 MBY71075.1 LRGB01000024 KZS21319.1 KT328074 ALB27752.1 ABLF02027371 ABLF02027373 GL732564 EFX77198.1 NEVH01020963 PNF20307.1 KF873587 AHJ11216.1 NWSH01001615 PCG70684.1 KZ150420 PZC70934.1 KP711055 AKQ12751.1 ADTU01018111 ADTU01018112 ADTU01018113 ADTU01018114 ADTU01018115 ADTU01018116 ADTU01018117 ADTU01018118 ADTU01018119 ADTU01018120 KRG03750.1 JX466888 AFU82559.1 KP711048 AKQ12744.1 EU082082 ABV45518.1 CH902624 KPU74285.1 JF731243 KP725465 AEA40429.1 AKV94622.1 CH940649 KRF81069.1 DQ498130 ABJ09669.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000218220
UP000001070
+ More
UP000030765 UP000001819 UP000007062 UP000007801 UP000000803 UP000008792 UP000002282 UP000008711 UP000192221 UP000183832 UP000007266 UP000007798 UP000037510 UP000000304 UP000009192 UP000001292 UP000037069 UP000092443 UP000268350 UP000192223 UP000076858 UP000095300 UP000007819 UP000000305 UP000235965 UP000075882 UP000005205
UP000030765 UP000001819 UP000007062 UP000007801 UP000000803 UP000008792 UP000002282 UP000008711 UP000192221 UP000183832 UP000007266 UP000007798 UP000037510 UP000000304 UP000009192 UP000001292 UP000037069 UP000092443 UP000268350 UP000192223 UP000076858 UP000095300 UP000007819 UP000000305 UP000235965 UP000075882 UP000005205
Interpro
Gene 3D
ProteinModelPortal
A8CGM5
A8HTC8
A0A0H4TRG2
A8HTC5
Q9XZI3
A0A0H4TQC6
+ More
A0A0B5GNX1 A0A194RGP8 A0A2H1W7E5 A0A212FIW5 A0A2A4K153 A0A2A4K033 A0A346IHV2 B4JD77 A0A1S4G7N3 A0A084VFG0 A0A1S4GZF8 Q29KA5 Q7PHY7 Q8T7V5 B3MMS1 A0A084VN01 A0A0J9R2A0 Q7KT97 B4M8Q7 B4P0I6 B3NA88 A0A1W4UXW1 Q7QI13 A0A1J1IWW5 A8DIU0 A8HTF2 B4MVM6 A9XFY4 A0A0L7LQI0 B4Q5H4 B4KEK2 B4HXE7 A0A0L0BUB3 A0A240SWJ8 E1JJR2 A0A0R1EB74 Q86MN7 B3NVM9 Q9VWI9 A0A3B0KMX8 B4PZJ0 B4MGC3 A4GXC7 A0A1W4VG42 B4L7L9 W8B2K3 A0A0C5I2P3 A0A348BSW2 B8X4G9 B4JJS4 A0A1W4X2L4 A0A348BSW4 A0A1Y1MEU7 A0A023GVG5 A0A348BSW3 A0A1D8GS61 G9CU47 K4JEQ6 A0A0R3P102 A0A3B0KA18 A0A0P5EP72 A0A2S2PZX2 A0A162CYS9 A0A0U2GQ98 A0A1I8PTD7 J9JT17 E9GTK4 A0A2J7PVE3 A0A1I8PTI3 W6EL40 A0A1I8PNY4 A0A182KMG8 A0A2A4JGH6 A0A2W1BAC7 A0A0H4TMF2 A0A158NJP1 A0A0Q9X676 K4JF52 A0A0H4U7U0 A8CGL6 A0A1I8NYF7 A0A0P8ZHV2 G9CU48 A0A0Q9W8H3 A5H032
A0A0B5GNX1 A0A194RGP8 A0A2H1W7E5 A0A212FIW5 A0A2A4K153 A0A2A4K033 A0A346IHV2 B4JD77 A0A1S4G7N3 A0A084VFG0 A0A1S4GZF8 Q29KA5 Q7PHY7 Q8T7V5 B3MMS1 A0A084VN01 A0A0J9R2A0 Q7KT97 B4M8Q7 B4P0I6 B3NA88 A0A1W4UXW1 Q7QI13 A0A1J1IWW5 A8DIU0 A8HTF2 B4MVM6 A9XFY4 A0A0L7LQI0 B4Q5H4 B4KEK2 B4HXE7 A0A0L0BUB3 A0A240SWJ8 E1JJR2 A0A0R1EB74 Q86MN7 B3NVM9 Q9VWI9 A0A3B0KMX8 B4PZJ0 B4MGC3 A4GXC7 A0A1W4VG42 B4L7L9 W8B2K3 A0A0C5I2P3 A0A348BSW2 B8X4G9 B4JJS4 A0A1W4X2L4 A0A348BSW4 A0A1Y1MEU7 A0A023GVG5 A0A348BSW3 A0A1D8GS61 G9CU47 K4JEQ6 A0A0R3P102 A0A3B0KA18 A0A0P5EP72 A0A2S2PZX2 A0A162CYS9 A0A0U2GQ98 A0A1I8PTD7 J9JT17 E9GTK4 A0A2J7PVE3 A0A1I8PTI3 W6EL40 A0A1I8PNY4 A0A182KMG8 A0A2A4JGH6 A0A2W1BAC7 A0A0H4TMF2 A0A158NJP1 A0A0Q9X676 K4JF52 A0A0H4U7U0 A8CGL6 A0A1I8NYF7 A0A0P8ZHV2 G9CU48 A0A0Q9W8H3 A5H032
PDB
4BOT
E-value=1.34621e-84,
Score=799
Ontologies
GO
GO:0030054
GO:0045211
GO:0016021
GO:0004888
GO:0022848
GO:0005887
GO:0045202
GO:0043005
GO:0007165
GO:0042391
GO:0007268
GO:0034220
GO:0050877
GO:0042166
GO:0005892
GO:0006812
GO:0005230
GO:0060079
GO:0007632
GO:0014069
GO:0030425
GO:0043197
GO:0007630
GO:0007271
GO:0006811
GO:0016020
GO:0006810
GO:0004889
GO:0005216
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
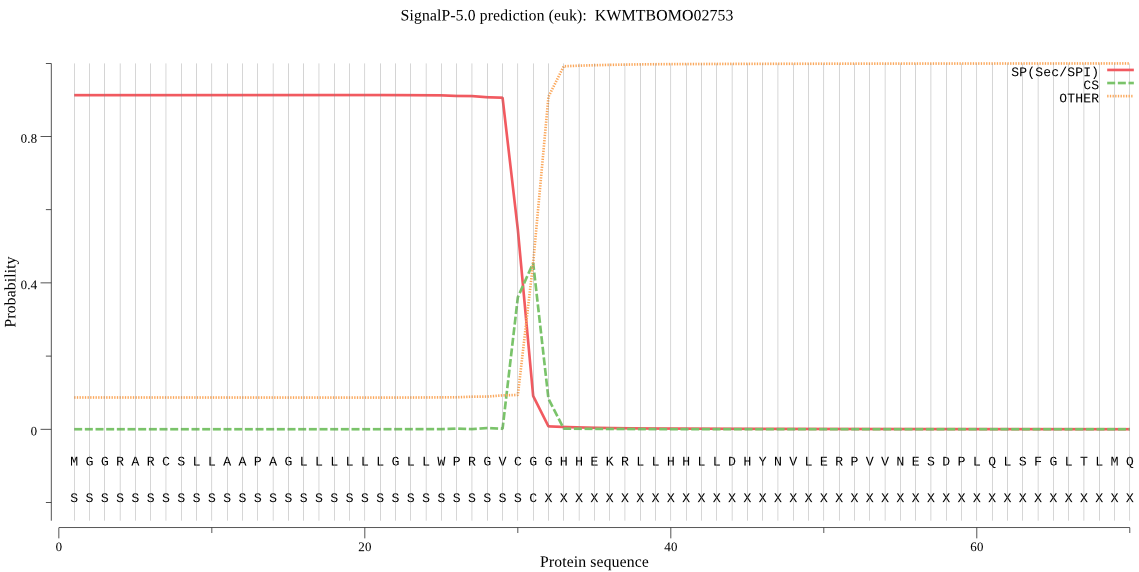
SignalP
Position: 1 - 31,
Likelihood: 0.913090
Length:
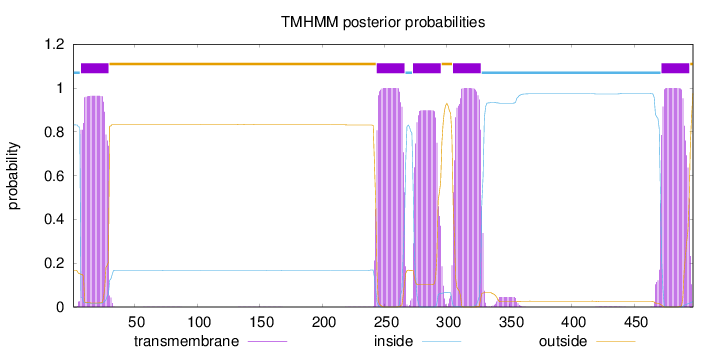
497
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
108.09138
Exp number, first 60 AAs:
21.18621
Total prob of N-in:
0.83224
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 243
TMhelix
244 - 266
inside
267 - 272
TMhelix
273 - 295
outside
296 - 304
TMhelix
305 - 327
inside
328 - 471
TMhelix
472 - 494
outside
495 - 497
Population Genetic Test Statistics
Pi
219.67232
Theta
178.06205
Tajima's D
1.502576
CLR
0.114315
CSRT
0.786260686965652
Interpretation
Uncertain
