Gene
KWMTBOMO02752
Pre Gene Modal
BGIBMGA003716
Annotation
nicotinic_acetylcholine_receptor_subunit_beta_1_isoform_1_precursor_[Bombyx_mori]
Full name
Acetylcholine receptor subunit beta-like 1
Alternative Name
Nicotinic acetylcholine receptor beta 1
Location in the cell
PlasmaMembrane Reliability : 4.8
Sequence
CDS
ATGTACCCGCATCCGGCAGTGCCCGCTGCAATGTCGTGCGTATCGCGCGCTTTCCTCGCCACCACCCTACTGGCCATCCTGTATTCAGGATGGTGTTCAGAAGACGAAGAACGTTTAGTTCGCGACCTTTTCAGAGGATACAATAAATTAATACGACCTGTCCAAAACATGACACAGAAAGTGGATGTGAGATTTGGTCTTGCGTTTGTACAACTCATTAACGTCAATGAGAAGAACCAAATTATGAAATCAAACGTTTGGCTCCGTTTGGTTTGGATGGATTACCAGTTGATGTGGGACGAAGCTGACTACGGCGGTATTGGCGTATTAAGATTACCGCCTGACAAGGTTTGGAAGCCAGATATAGTTCTTTTCAATAACGCTGACGGAAATTACGAAGTCCGATATAAATCAAACGTGCTTATATATCCTAATGGAGAAGTACTTTGGGTCCCACCAGCTATATATCAGAGCTCGTGCACGATTGACGTAACCTACTTTCCATTTGACCAGCAAACGTGTATCATGAAATTCGGATCTTGGACGTTCAATGGGGACCAAGTATCACTGGCGCTATACAACAACAAGAACTTTGTTGATCTTTCCGATTATTGGAAGTCAGGAACATGGGATATCATAGAAGTACCGGCTTATTTGAACATTTATGAAGGCAATCATCCAACTGAAACTGATATTACGTTCTACATTATAATTCGACGGAAAACATTGTTTTACACTGTAAACTTAATATTGCCGACTGTTTTAATTTCGTTTCTTTGTGTACTTGTATTTTATTTGCCAGCTGAAGCTGGTGAAAAGGTAACCTTGGGTATCAGCATTTTACTTTCTCTGGTTGTGTTCCTGCTTCTGGTATCGAAGATTCTTCCACCGACATCGCTTGTACTTCCACTTATCGCTAAATATTTATTATTCACATTCATTATGAATACTGTCAGTATTCTGGTTACGGTTATCATAATCAATTGGAACTTCAGAGGACCCAGAACTCATAGGATGCCATTATGGATTCGAAGCGTTTTCCTGCATTATTTACCAGCTATGCTGCTTATGCGGAGACCGAGAAAAACTAGATTGCGCTGGATGATGGAAATGCCCGGAATGGGTGCTCCACCCCATGCAGCGGCACCACACGACCTCCCGAAACATATAAGCAAAATGGAGGCAATGGAGCTATCGGACCTACATCATCCAAATTGCAAAATAAACCGTGCTGCCGGCGGAGGTGAAGTTGGCGCTCTTGGAGATCTTGGTGCACTGGGTGGTCTGGGACTTGGAGGAGAAAGACGCGAATCGGAAAGTTCGGATTCACTTCTACTTTCTCCTGAAGCGGCAAAGGCTACGGAAGCAGTCGAATTCATTGCGGAACATTTACGAAATGAAGATCTTTACATTCAGACTCGCGAAGACTGGAAATACGTGGCAATGGTTATTGACAGATTACAGCTGTACATTTTCTTCATAGTCACCACAGCAGGAACGGTTGGGATACTGATGGATGCACCACATATTTTCGAATATGTTGACCAAGATCGTATTATTGAAATTTATCGAGGAAAATAA
Protein
MYPHPAVPAAMSCVSRAFLATTLLAILYSGWCSEDEERLVRDLFRGYNKLIRPVQNMTQKVDVRFGLAFVQLINVNEKNQIMKSNVWLRLVWMDYQLMWDEADYGGIGVLRLPPDKVWKPDIVLFNNADGNYEVRYKSNVLIYPNGEVLWVPPAIYQSSCTIDVTYFPFDQQTCIMKFGSWTFNGDQVSLALYNNKNFVDLSDYWKSGTWDIIEVPAYLNIYEGNHPTETDITFYIIIRRKTLFYTVNLILPTVLISFLCVLVFYLPAEAGEKVTLGISILLSLVVFLLLVSKILPPTSLVLPLIAKYLLFTFIMNTVSILVTVIIINWNFRGPRTHRMPLWIRSVFLHYLPAMLLMRRPRKTRLRWMMEMPGMGAPPHAAAPHDLPKHISKMEAMELSDLHHPNCKINRAAGGGEVGALGDLGALGGLGLGGERRESESSDSLLLSPEAAKATEAVEFIAEHLRNEDLYIQTREDWKYVAMVIDRLQLYIFFIVTTAGTVGILMDAPHIFEYVDQDRIIEIYRGK
Summary
Description
After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane.
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Keywords
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Polymorphism
Postsynaptic cell membrane
Receptor
Reference proteome
Signal
Synapse
Transmembrane
Transmembrane helix
Transport
Feature
chain Acetylcholine receptor subunit beta-like 1
Uniprot
A8HTD6
A8CGJ8
A0A2H1W6U0
A0A212FIW5
O96633
V5ULZ2
+ More
A0A2A4K0T0 A0A346IHV5 A0A194RLA2 A7UHB4 A0A0H4TM39 A0A182RH67 A0A1B0FB86 W5JBF0 A0A182PPX6 A0A182MEW2 A0A182V2W0 A0A182TNK2 A0A182XNI3 Q7QI10 A0A182MZW4 A0A182QXM9 A0A182IJQ9 A0A084VN07 A0A182W452 A8DIV7 D7EIM1 A0A182IDN6 A0A1A9V495 A0A1A9W4T0 A0A182YRJ4 A0A1Y1NKI0 A0A1B0DLT1 A0A3B0J6I3 C3TS68 B4KX76 Q16SL2 A0A1W4VAZ3 B4MN11 B4LGV9 A0A1J1HTP9 B0X9Y9 A0A0J9RQG6 A0A0Q9XAQ7 A0A3B0K2J2 B4PI73 B3NC38 E8NHB5 A0A0Q5U785 P04755 A0A0K8UGV2 A0A0K1U4A8 Q29EC6 B4HU63 A0A348BSW8 B3M928 A0A2H4U2F6 A0A034VYY8 W8ARB5 A0A2Z4N5Q8 B6VAH8 A0A0G3VJ88 A0A0A1X5B9 A0A0P8ZTC8 A0A0J7P1B2 A0A195E3M5 Q30DN7 A0A1I8M6L9 A0A1W4X3R6 A0A195D2I7 W6EL29 A0A154P0V3 A0A3L8DSH3 A0A1I8PPB4 T1PA71 A0A158P1F1 F6JX92 A0EIZ8 A0A151I0X7 A0A0C9QPC9 A0A195EXD0 H9J2H9 A0A182FIN7 O46135 A0A310SUJ8 E0VP62 T1IAD6 A0A151XHB4 A0A0A7RQC5 D3UA25 A0A1D8GS81 F4WV01 A0A1S5VZ00 A0A1B0B7V4 J9QIC9 A0A0U2GT18 A0A224AUJ9 A0A2S2N7R3 Q9NFX8 J9K5U1
A0A2A4K0T0 A0A346IHV5 A0A194RLA2 A7UHB4 A0A0H4TM39 A0A182RH67 A0A1B0FB86 W5JBF0 A0A182PPX6 A0A182MEW2 A0A182V2W0 A0A182TNK2 A0A182XNI3 Q7QI10 A0A182MZW4 A0A182QXM9 A0A182IJQ9 A0A084VN07 A0A182W452 A8DIV7 D7EIM1 A0A182IDN6 A0A1A9V495 A0A1A9W4T0 A0A182YRJ4 A0A1Y1NKI0 A0A1B0DLT1 A0A3B0J6I3 C3TS68 B4KX76 Q16SL2 A0A1W4VAZ3 B4MN11 B4LGV9 A0A1J1HTP9 B0X9Y9 A0A0J9RQG6 A0A0Q9XAQ7 A0A3B0K2J2 B4PI73 B3NC38 E8NHB5 A0A0Q5U785 P04755 A0A0K8UGV2 A0A0K1U4A8 Q29EC6 B4HU63 A0A348BSW8 B3M928 A0A2H4U2F6 A0A034VYY8 W8ARB5 A0A2Z4N5Q8 B6VAH8 A0A0G3VJ88 A0A0A1X5B9 A0A0P8ZTC8 A0A0J7P1B2 A0A195E3M5 Q30DN7 A0A1I8M6L9 A0A1W4X3R6 A0A195D2I7 W6EL29 A0A154P0V3 A0A3L8DSH3 A0A1I8PPB4 T1PA71 A0A158P1F1 F6JX92 A0EIZ8 A0A151I0X7 A0A0C9QPC9 A0A195EXD0 H9J2H9 A0A182FIN7 O46135 A0A310SUJ8 E0VP62 T1IAD6 A0A151XHB4 A0A0A7RQC5 D3UA25 A0A1D8GS81 F4WV01 A0A1S5VZ00 A0A1B0B7V4 J9QIC9 A0A0U2GT18 A0A224AUJ9 A0A2S2N7R3 Q9NFX8 J9K5U1
Pubmed
17868469
22118469
26354079
20920257
23761445
12364791
+ More
14747013 15676276 17210077 24438588 17880682 18362917 19820115 25244985 28004739 19320762 17994087 17510324 22936249 17550304 16453690 3136037 2832736 10731132 12537572 15632085 18057021 25348373 24495485 19254762 25830018 25315136 25701599 30249741 21347285 21618599 17065616 19121390 9660807 20566863 25504620 20087392 20075255 21719571 28187830
14747013 15676276 17210077 24438588 17880682 18362917 19820115 25244985 28004739 19320762 17994087 17510324 22936249 17550304 16453690 3136037 2832736 10731132 12537572 15632085 18057021 25348373 24495485 19254762 25830018 25315136 25701599 30249741 21347285 21618599 17065616 19121390 9660807 20566863 25504620 20087392 20075255 21719571 28187830
EMBL
EU143801
ABV72692.1
EU082071
ABV45508.1
ODYU01006685
SOQ48753.1
+ More
AGBW02008321 OWR53672.1 AF096880 AAD09810.1 KF668454 AHB78808.1 NWSH01000300 PCG77626.1 MH138169 AXP17110.1 KQ460207 KPJ16761.1 EU070937 EU101456 ABU41521.1 ABU85033.1 KP711052 RSAL01000017 AKQ12748.1 RVE52947.1 CCAG010009347 ADMH02001869 ETN60728.1 AXCM01005037 AY705405 AAAB01008811 AAU12514.1 EAA04968.3 AXCN02002024 ATLV01014666 KE524978 KFB39351.1 EF526094 ABS86916.1 KQ971307 EFA12056.2 GEZM01000477 JAV98401.1 AJVK01006807 AJVK01006808 OUUW01000002 SPP77607.1 EU937811 ACP31316.1 CH933809 EDW17534.1 CH477672 EAT37452.1 CH963847 EDW73567.1 CH940647 EDW68429.1 CVRI01000014 CRK89854.1 DS232556 EDS43415.1 CM002912 KMY97609.1 KRG05606.1 SPP77608.1 CM000159 EDW93415.1 CH954178 EDV50926.1 BT125991 ADX35970.1 KQS43588.1 X04016 X07956 X07957 X07958 M20316 AE014296 GDHF01026526 JAI25788.1 KP725467 AKV94624.1 CH379070 EAL30135.2 CH480817 EDW50484.1 LC389192 BBE49571.1 CH902618 EDV38972.1 KPU77787.1 KY293419 ATZ35994.1 GAKP01012214 JAC46738.1 GAMC01015165 JAB91390.1 MF612140 AWX65628.1 FJ358493 ACJ07013.1 KP339870 AKL79440.1 GBXI01008467 GBXI01008196 GBXI01000170 JAD05825.1 JAD06096.1 JAD14122.1 KPU77786.1 LBMM01000399 KMQ98445.1 KQ979709 KYN19474.1 DQ153040 ABA39253.1 KQ976986 KYN06589.1 KF873577 AHJ11206.1 KQ434796 KZC05556.1 QOIP01000004 RLU23395.1 KA645589 AFP60218.1 ADTU01006423 ADTU01006424 HM049634 HM049635 ADH15832.1 DQ026038 KJ939597 AAY87897.1 AJE70268.1 KQ976633 KYM78882.1 GBYB01002547 JAG72314.1 KQ981928 KYN32883.1 BABH01007978 AJ000393 CAA04055.1 KQ759901 OAD62012.1 DS235356 EEB15168.1 ACPB03019553 KQ982138 KYQ59600.1 KP101249 AJA39825.1 FJ821444 ACY82698.1 KU557782 AOT81844.1 GL888382 EGI61968.1 KX602254 AQP31213.1 JXJN01009725 JXJN01009726 JXJN01009727 JN681174 AFP55243.1 KT328076 ALB27754.1 LC215870 BBA21165.1 GGMR01000632 MBY13251.1 AJ251838 CAB87995.1 ABLF02038405
AGBW02008321 OWR53672.1 AF096880 AAD09810.1 KF668454 AHB78808.1 NWSH01000300 PCG77626.1 MH138169 AXP17110.1 KQ460207 KPJ16761.1 EU070937 EU101456 ABU41521.1 ABU85033.1 KP711052 RSAL01000017 AKQ12748.1 RVE52947.1 CCAG010009347 ADMH02001869 ETN60728.1 AXCM01005037 AY705405 AAAB01008811 AAU12514.1 EAA04968.3 AXCN02002024 ATLV01014666 KE524978 KFB39351.1 EF526094 ABS86916.1 KQ971307 EFA12056.2 GEZM01000477 JAV98401.1 AJVK01006807 AJVK01006808 OUUW01000002 SPP77607.1 EU937811 ACP31316.1 CH933809 EDW17534.1 CH477672 EAT37452.1 CH963847 EDW73567.1 CH940647 EDW68429.1 CVRI01000014 CRK89854.1 DS232556 EDS43415.1 CM002912 KMY97609.1 KRG05606.1 SPP77608.1 CM000159 EDW93415.1 CH954178 EDV50926.1 BT125991 ADX35970.1 KQS43588.1 X04016 X07956 X07957 X07958 M20316 AE014296 GDHF01026526 JAI25788.1 KP725467 AKV94624.1 CH379070 EAL30135.2 CH480817 EDW50484.1 LC389192 BBE49571.1 CH902618 EDV38972.1 KPU77787.1 KY293419 ATZ35994.1 GAKP01012214 JAC46738.1 GAMC01015165 JAB91390.1 MF612140 AWX65628.1 FJ358493 ACJ07013.1 KP339870 AKL79440.1 GBXI01008467 GBXI01008196 GBXI01000170 JAD05825.1 JAD06096.1 JAD14122.1 KPU77786.1 LBMM01000399 KMQ98445.1 KQ979709 KYN19474.1 DQ153040 ABA39253.1 KQ976986 KYN06589.1 KF873577 AHJ11206.1 KQ434796 KZC05556.1 QOIP01000004 RLU23395.1 KA645589 AFP60218.1 ADTU01006423 ADTU01006424 HM049634 HM049635 ADH15832.1 DQ026038 KJ939597 AAY87897.1 AJE70268.1 KQ976633 KYM78882.1 GBYB01002547 JAG72314.1 KQ981928 KYN32883.1 BABH01007978 AJ000393 CAA04055.1 KQ759901 OAD62012.1 DS235356 EEB15168.1 ACPB03019553 KQ982138 KYQ59600.1 KP101249 AJA39825.1 FJ821444 ACY82698.1 KU557782 AOT81844.1 GL888382 EGI61968.1 KX602254 AQP31213.1 JXJN01009725 JXJN01009726 JXJN01009727 JN681174 AFP55243.1 KT328076 ALB27754.1 LC215870 BBA21165.1 GGMR01000632 MBY13251.1 AJ251838 CAB87995.1 ABLF02038405
Proteomes
UP000007151
UP000218220
UP000053240
UP000283053
UP000075900
UP000092444
+ More
UP000000673 UP000075885 UP000075883 UP000075903 UP000075902 UP000076407 UP000007062 UP000075884 UP000075886 UP000075880 UP000030765 UP000075920 UP000007266 UP000078200 UP000091820 UP000076408 UP000092462 UP000268350 UP000009192 UP000008820 UP000192221 UP000007798 UP000008792 UP000183832 UP000002320 UP000002282 UP000008711 UP000000803 UP000001819 UP000001292 UP000007801 UP000036403 UP000078492 UP000095301 UP000192223 UP000078542 UP000076502 UP000279307 UP000095300 UP000005205 UP000005203 UP000078540 UP000078541 UP000005204 UP000069272 UP000009046 UP000015103 UP000075809 UP000002358 UP000007755 UP000092460 UP000007819
UP000000673 UP000075885 UP000075883 UP000075903 UP000075902 UP000076407 UP000007062 UP000075884 UP000075886 UP000075880 UP000030765 UP000075920 UP000007266 UP000078200 UP000091820 UP000076408 UP000092462 UP000268350 UP000009192 UP000008820 UP000192221 UP000007798 UP000008792 UP000183832 UP000002320 UP000002282 UP000008711 UP000000803 UP000001819 UP000001292 UP000007801 UP000036403 UP000078492 UP000095301 UP000192223 UP000078542 UP000076502 UP000279307 UP000095300 UP000005205 UP000005203 UP000078540 UP000078541 UP000005204 UP000069272 UP000009046 UP000015103 UP000075809 UP000002358 UP000007755 UP000092460 UP000007819
Interpro
Gene 3D
ProteinModelPortal
A8HTD6
A8CGJ8
A0A2H1W6U0
A0A212FIW5
O96633
V5ULZ2
+ More
A0A2A4K0T0 A0A346IHV5 A0A194RLA2 A7UHB4 A0A0H4TM39 A0A182RH67 A0A1B0FB86 W5JBF0 A0A182PPX6 A0A182MEW2 A0A182V2W0 A0A182TNK2 A0A182XNI3 Q7QI10 A0A182MZW4 A0A182QXM9 A0A182IJQ9 A0A084VN07 A0A182W452 A8DIV7 D7EIM1 A0A182IDN6 A0A1A9V495 A0A1A9W4T0 A0A182YRJ4 A0A1Y1NKI0 A0A1B0DLT1 A0A3B0J6I3 C3TS68 B4KX76 Q16SL2 A0A1W4VAZ3 B4MN11 B4LGV9 A0A1J1HTP9 B0X9Y9 A0A0J9RQG6 A0A0Q9XAQ7 A0A3B0K2J2 B4PI73 B3NC38 E8NHB5 A0A0Q5U785 P04755 A0A0K8UGV2 A0A0K1U4A8 Q29EC6 B4HU63 A0A348BSW8 B3M928 A0A2H4U2F6 A0A034VYY8 W8ARB5 A0A2Z4N5Q8 B6VAH8 A0A0G3VJ88 A0A0A1X5B9 A0A0P8ZTC8 A0A0J7P1B2 A0A195E3M5 Q30DN7 A0A1I8M6L9 A0A1W4X3R6 A0A195D2I7 W6EL29 A0A154P0V3 A0A3L8DSH3 A0A1I8PPB4 T1PA71 A0A158P1F1 F6JX92 A0EIZ8 A0A151I0X7 A0A0C9QPC9 A0A195EXD0 H9J2H9 A0A182FIN7 O46135 A0A310SUJ8 E0VP62 T1IAD6 A0A151XHB4 A0A0A7RQC5 D3UA25 A0A1D8GS81 F4WV01 A0A1S5VZ00 A0A1B0B7V4 J9QIC9 A0A0U2GT18 A0A224AUJ9 A0A2S2N7R3 Q9NFX8 J9K5U1
A0A2A4K0T0 A0A346IHV5 A0A194RLA2 A7UHB4 A0A0H4TM39 A0A182RH67 A0A1B0FB86 W5JBF0 A0A182PPX6 A0A182MEW2 A0A182V2W0 A0A182TNK2 A0A182XNI3 Q7QI10 A0A182MZW4 A0A182QXM9 A0A182IJQ9 A0A084VN07 A0A182W452 A8DIV7 D7EIM1 A0A182IDN6 A0A1A9V495 A0A1A9W4T0 A0A182YRJ4 A0A1Y1NKI0 A0A1B0DLT1 A0A3B0J6I3 C3TS68 B4KX76 Q16SL2 A0A1W4VAZ3 B4MN11 B4LGV9 A0A1J1HTP9 B0X9Y9 A0A0J9RQG6 A0A0Q9XAQ7 A0A3B0K2J2 B4PI73 B3NC38 E8NHB5 A0A0Q5U785 P04755 A0A0K8UGV2 A0A0K1U4A8 Q29EC6 B4HU63 A0A348BSW8 B3M928 A0A2H4U2F6 A0A034VYY8 W8ARB5 A0A2Z4N5Q8 B6VAH8 A0A0G3VJ88 A0A0A1X5B9 A0A0P8ZTC8 A0A0J7P1B2 A0A195E3M5 Q30DN7 A0A1I8M6L9 A0A1W4X3R6 A0A195D2I7 W6EL29 A0A154P0V3 A0A3L8DSH3 A0A1I8PPB4 T1PA71 A0A158P1F1 F6JX92 A0EIZ8 A0A151I0X7 A0A0C9QPC9 A0A195EXD0 H9J2H9 A0A182FIN7 O46135 A0A310SUJ8 E0VP62 T1IAD6 A0A151XHB4 A0A0A7RQC5 D3UA25 A0A1D8GS81 F4WV01 A0A1S5VZ00 A0A1B0B7V4 J9QIC9 A0A0U2GT18 A0A224AUJ9 A0A2S2N7R3 Q9NFX8 J9K5U1
PDB
6CNK
E-value=2.06552e-91,
Score=858
Ontologies
GO
PANTHER
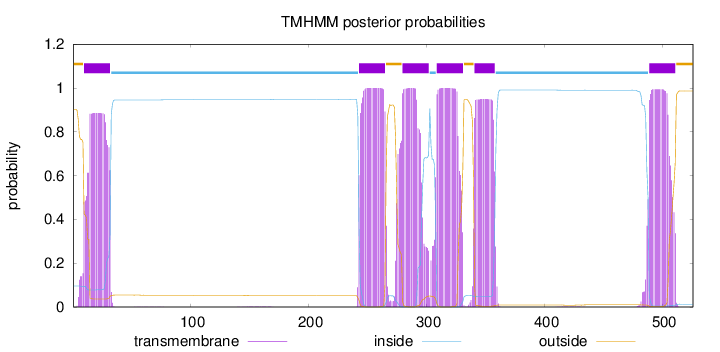
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
Length:
526
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
124.06553
Exp number, first 60 AAs:
18.59015
Total prob of N-in:
0.09647
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 242
TMhelix
243 - 265
outside
266 - 279
TMhelix
280 - 302
inside
303 - 308
TMhelix
309 - 331
outside
332 - 340
TMhelix
341 - 358
inside
359 - 488
TMhelix
489 - 511
outside
512 - 526
Population Genetic Test Statistics
Pi
207.846011
Theta
170.306126
Tajima's D
0.913785
CLR
0.46917
CSRT
0.635818209089546
Interpretation
Uncertain
