Gene
KWMTBOMO02749 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003717
Annotation
PREDICTED:_ATP-dependent_6-phosphofructokinase_isoform_X1_[Bombyx_mori]
Full name
ATP-dependent 6-phosphofructokinase
Alternative Name
Phosphohexokinase
Location in the cell
Cytoplasmic Reliability : 1.764
Sequence
CDS
ATGGACGGGTCAGCTACTCAGAAATTTATTGGCCGCGGCTCTCATAAAGGCAAGGGGCTTGCTGTGTTTACTAGCGGTGGAGATTCTCAGGGAATGAATGCCGCTGTGCGATCAGTCGTGCGTATGGGCATATACTTAGGGTGTAAAGTATATTTTATTCGTGAAGGATACCAGGGCATGGTAGATGGAGGTGATAACATTGAAGAAGCGAACTGGTCTTCTGTTAGCTCTATAATTCACAAAGGAGGCACTATAATTGGATCAGCTAGGTGCATGGAGTTCATTAAAAAAGAAGGTCGTCTCAAAGCCGCATATAACTTAGTGAGCCGTGGTATAACCAATCTTGTGGTTATTGGTGGTGATGGTTCGCTGACTGGTGCCAATTTGTTCCGACAAGAATGGTCTAGCCTTCTTGATGAATTGCTGGAGAACAACAAAATTACTAAAGATCAAAGGGAAAAATATAAATTTTTGCACATAGCTGGAATGGTTGGTTCAATTGACAATGATTTTTGTGGTACAGACATGACAATAGGTACTGATTCAGCCTTGCATAGAATCATTGAAGCTATTGATGCAATTGTAAGCACTGCTTATTCACATCAAAGAACATTTATTATGGAGGTCATGGGTCGCAACTGTGGTTATTTGGCCCTTGTCGCTGCTCTGGCGAGCGAGGCAGACCAAGTATTTATACCTGAAGACCCTGTTCCCAATAATTGGGTTGAAAAACTTTGCAAAAGATTGCAACAGGAGAGGCGTTCTGGTCAACGTTTGAATATTATCATTGTGGCCGAGGGTGCGATCGACCGCGAAGGTAAGCCTATCACCGCCGAGTTGGTGAAGAAGGTGGTGGTTGATAACCTGCAGCAGGATACCCGTATTACCGTGTTGGGTCACGTCCAACGAGGAGGATCTCCCTCCGCCTTCGACAGAATTTTGGGATGTCGTATGGGAGCAGAGGCGGTTATGGCTTTAATGGAGGCCACCCCGGAAACCGAACCGTGTGTTGTTTCGCTCGACGGTAACCAAGCTGTTCGTTTGCCGTTAATGGAATGCGTTCGACGAACGAAAGCTGTCTCACAGGCAATGGCTGATAAAAATTGGGACCTAGCTGTACAGCTCCGCGGTCGTAGTTTTGCACGTAATTTGGAGACATATAAGATGTTGACCCGTTTGAAGCCACCAAAGGGAGCGTTTGATGAATCCGGAAAACCTTCTGAAGGTTTGACGCTGGCCGTGATGCATGTGGGCGCGCCGGCGTGCGGCATGAATGCGGCCGTACGTTCGTTTGTACGGAATTGTATTTACCGAGGCGATACCGTCCTTGGCATTCACGACGGCATTGAAGGTTTCATTAACGGGAACATCGTCCAAATGGAATGGTCGGATGTAACGGGTTGGGTTGCTCAAGGAGGCGCTTTTCTTGGTACAAAGAGGACTTTGCCCGGCAATAGGAAGAGTGACATCGCTGCGCGGATAAAGCAGTTCAATATTCAAGGCTTACTCATCATCGGTGGTTTTGAGGCCTACCAAGCTAGTTTGGAACTATACGAATCTCGCGCTGAATTCCCGGAGTTCTGCATTCCTCTGGTCGTCATCCCGTCCACGATAAGTAACAACGTGCCCGGGACGGATTTCTCTCTGGGAGCCGACACTGCGCTCAATGAAATAACTGAGATCTGCGATCGCATTCGTCAGTCGGCACAGGGTACCAAGCGTCGTGTGTTCGTCATCGAAACCATGGGCGGTTACTGCGGTTACCTCGCTACCTTGGCTGGTTTGGCCGGCGGTGCTGATGCCGCATACATTTACGAGGAGAAGTTCTCCATCAAGGATCTGCAACAGGACGTTTATCACATGGCGTCTAAAATGACCGGCGGCATTCAGCGCGGCCTCATCCTGCGCAATGAGAAAGCGAACGATAACTACAACACGGAGTTCATTTACCGACTGTACTCTGAAGAGGGCAAAGGACTGTTTACAGCTCGCATGAATGTGCTCGGTCACATGCAACAGGGCGGCTCTCCGACGCCATTTGATCGTAACATGGGTACTAAGATGGCGGCCAAATGTCTTCATTGGCTCGTTGAGAAAATTCAGGAAGGTCCCTCGAACTCTGCCGACTCTGCTTGTTTACTTGGTGTTGTTAAACGCCAGTACAAATTCACTCCACTCGAAGAACTCAAAACTCAAACTAATTTTGCTCAAAGAATAGCAAAACAGCAATGGTGGATGAAACTGCGTCCATTGCTTCGTATCCTGGCGAAGCACGATTCTACATACGAAGAAGAAGGAATGTATATGACAGTTGAACAAGGAGAAATCGACTCTGAGAACGTTATTTAA
Protein
MDGSATQKFIGRGSHKGKGLAVFTSGGDSQGMNAAVRSVVRMGIYLGCKVYFIREGYQGMVDGGDNIEEANWSSVSSIIHKGGTIIGSARCMEFIKKEGRLKAAYNLVSRGITNLVVIGGDGSLTGANLFRQEWSSLLDELLENNKITKDQREKYKFLHIAGMVGSIDNDFCGTDMTIGTDSALHRIIEAIDAIVSTAYSHQRTFIMEVMGRNCGYLALVAALASEADQVFIPEDPVPNNWVEKLCKRLQQERRSGQRLNIIIVAEGAIDREGKPITAELVKKVVVDNLQQDTRITVLGHVQRGGSPSAFDRILGCRMGAEAVMALMEATPETEPCVVSLDGNQAVRLPLMECVRRTKAVSQAMADKNWDLAVQLRGRSFARNLETYKMLTRLKPPKGAFDESGKPSEGLTLAVMHVGAPACGMNAAVRSFVRNCIYRGDTVLGIHDGIEGFINGNIVQMEWSDVTGWVAQGGAFLGTKRTLPGNRKSDIAARIKQFNIQGLLIIGGFEAYQASLELYESRAEFPEFCIPLVVIPSTISNNVPGTDFSLGADTALNEITEICDRIRQSAQGTKRRVFVIETMGGYCGYLATLAGLAGGADAAYIYEEKFSIKDLQQDVYHMASKMTGGIQRGLILRNEKANDNYNTEFIYRLYSEEGKGLFTARMNVLGHMQQGGSPTPFDRNMGTKMAAKCLHWLVEKIQEGPSNSADSACLLGVVKRQYKFTPLEELKTQTNFAQRIAKQQWWMKLRPLLRILAKHDSTYEEEGMYMTVEQGEIDSENVI
Summary
Description
Catalyzes the phosphorylation of D-fructose 6-phosphate to fructose 1,6-bisphosphate by ATP, the first committing step of glycolysis.
Catalytic Activity
ATP + beta-D-fructose 6-phosphate = ADP + beta-D-fructose 1,6-bisphosphate + H(+)
Cofactor
Mg(2+)
Subunit
Homotetramer.
Similarity
Belongs to the phosphofructokinase type A (PFKA) family. ATP-dependent PFK group I subfamily. Eukaryotic two domain clade "E" sub-subfamily.
Keywords
Allosteric enzyme
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Glycolysis
Kinase
Magnesium
Metal-binding
Nucleotide-binding
Reference proteome
Transferase
Feature
chain ATP-dependent 6-phosphofructokinase
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9J2I0
A0A2H1W9K4
A0A194PQ53
A0A0F6TNF4
A0A194RH42
A0A2A4K1T8
+ More
A0A212FIX8 S4NSV3 A0A2M4BEL6 A0A182FIA4 A0A2M4A324 A0A2M4A375 W5JTT0 A0A1C7CZ10 A0A2M4BEC6 A0A1Y9IXF3 A0A1Y9IRG4 A0A182I3Q9 A0A1Y9I2A6 A0A2M4BED7 A0A182XZI2 A0A1Q3FML9 A0A2M3Z2T4 A0A2M4A337 A0A240PN31 A0A1Y9H0Y5 A0A2K6V9F9 A0A084VFB0 A0A2K6V9F7 A0A182RFP8 A0A023EV74 A0A182MUK8 A0A1B6CPV7 A0A1Y9H9G2 A0A1B6DHL7 A0A182WIS9 A0A1B6DAI8 A0A1Y9IXX5 A0A1Y9IRZ8 A0A1S4GVQ5 A0A1Y9GIZ8 A0A182KRV4 A0A1Y9IS69 A0A1Y9GJI9 A0A182X6F6 Q5TX32 A0A1Q3FMX3 A0A240PNR8 A0A240PPG1 A0A1Q3FMW2 U5EUA4 A0A182P3I4 J9JNX0 A0A1Y9H0J2 A0A1Y9HAX6 A0A182NHK2 A0A1L8E2A1 A0A1Y9HAX5 A0A2C9GUF9 A0A1Y9HA64 A0A1Y9HAY6 Q174J0 A0A2C9GUI7 A0A1Y9IUN1 A0A1Y9IUM9 A0A0K8TN59 A0A182J6F9 A0A240PMF4 A0A336LMI2 B0WNP9 A0A2H8TYE0 W8B915 A0A034V1J7 A0A067QZR7 A0A0K8VIR0 A0A2J7QM39 A0A1W5YLK2 A0A2Z5RE15 A0A0A1X4W6 E0W457 A0A1I8MFB5 V5GLY8 A0A1I8MFB9 A0A1I8MFC8 A0A0K8V1T2 A0A034V5U4 A0A0A1X492 T1PDK0 A0A1I8NRU4 A0A1I8NRU1 A0A1I8NRU9 A0A1B0B232 A0A1A9WMU0 A0A1A9VY37 A0A1B0AGU6 B4KSJ9 A0A1A9YNY9 A0A0J9R9B9 P52034-3 C7LA80
A0A212FIX8 S4NSV3 A0A2M4BEL6 A0A182FIA4 A0A2M4A324 A0A2M4A375 W5JTT0 A0A1C7CZ10 A0A2M4BEC6 A0A1Y9IXF3 A0A1Y9IRG4 A0A182I3Q9 A0A1Y9I2A6 A0A2M4BED7 A0A182XZI2 A0A1Q3FML9 A0A2M3Z2T4 A0A2M4A337 A0A240PN31 A0A1Y9H0Y5 A0A2K6V9F9 A0A084VFB0 A0A2K6V9F7 A0A182RFP8 A0A023EV74 A0A182MUK8 A0A1B6CPV7 A0A1Y9H9G2 A0A1B6DHL7 A0A182WIS9 A0A1B6DAI8 A0A1Y9IXX5 A0A1Y9IRZ8 A0A1S4GVQ5 A0A1Y9GIZ8 A0A182KRV4 A0A1Y9IS69 A0A1Y9GJI9 A0A182X6F6 Q5TX32 A0A1Q3FMX3 A0A240PNR8 A0A240PPG1 A0A1Q3FMW2 U5EUA4 A0A182P3I4 J9JNX0 A0A1Y9H0J2 A0A1Y9HAX6 A0A182NHK2 A0A1L8E2A1 A0A1Y9HAX5 A0A2C9GUF9 A0A1Y9HA64 A0A1Y9HAY6 Q174J0 A0A2C9GUI7 A0A1Y9IUN1 A0A1Y9IUM9 A0A0K8TN59 A0A182J6F9 A0A240PMF4 A0A336LMI2 B0WNP9 A0A2H8TYE0 W8B915 A0A034V1J7 A0A067QZR7 A0A0K8VIR0 A0A2J7QM39 A0A1W5YLK2 A0A2Z5RE15 A0A0A1X4W6 E0W457 A0A1I8MFB5 V5GLY8 A0A1I8MFB9 A0A1I8MFC8 A0A0K8V1T2 A0A034V5U4 A0A0A1X492 T1PDK0 A0A1I8NRU4 A0A1I8NRU1 A0A1I8NRU9 A0A1B0B232 A0A1A9WMU0 A0A1A9VY37 A0A1B0AGU6 B4KSJ9 A0A1A9YNY9 A0A0J9R9B9 P52034-3 C7LA80
EC Number
2.7.1.11
Pubmed
EMBL
BABH01007979
ODYU01007126
SOQ49626.1
KQ459597
KPI94869.1
KP253076
+ More
AKE50539.1 KQ460207 KPJ16759.1 NWSH01000300 PCG77630.1 AGBW02008321 OWR53675.1 GAIX01013897 JAA78663.1 GGFJ01002260 MBW51401.1 GGFK01001895 MBW35216.1 GGFK01001916 MBW35237.1 ADMH02000160 ETN67536.1 GGFJ01002259 MBW51400.1 APCN01000218 AAAB01008807 GGFJ01002261 MBW51402.1 GFDL01006186 JAV28859.1 GGFM01002081 MBW22832.1 GGFK01001902 MBW35223.1 ATLV01012372 KE524785 KFB36654.1 JXUM01042935 JXUM01042936 JXUM01042937 GAPW01000248 KQ561343 JAC13350.1 KXJ78899.1 AXCM01004236 GEDC01022015 JAS15283.1 AXCN02000593 GEDC01020516 GEDC01012156 JAS16782.1 JAS25142.1 GEDC01014581 GEDC01003943 JAS22717.1 JAS33355.1 EAL41738.3 GFDL01006222 JAV28823.1 GFDL01006232 JAV28813.1 GANO01001572 JAB58299.1 ABLF02041209 ABLF02041211 GFDF01001372 JAV12712.1 CH477410 EAT41468.1 GDAI01001796 JAI15807.1 UFQT01000063 SSX19272.1 DS232014 EDS31890.1 GFXV01007518 MBW19323.1 GAMC01016889 JAB89666.1 GAKP01021821 JAC37131.1 KK852854 KDR15026.1 GDHF01013540 JAI38774.1 NEVH01013227 PNF29623.1 KX147660 ARI45064.1 FX983726 BAX07225.1 GBXI01011096 GBXI01008564 JAD03196.1 JAD05728.1 DS235886 EEB20413.1 GALX01003377 JAB65089.1 GDHF01033073 GDHF01019749 JAI19241.1 JAI32565.1 GAKP01021822 JAC37130.1 GBXI01008380 JAD05912.1 KA646210 AFP60839.1 JXJN01007398 JXJN01007399 CH933808 EDW09504.2 CM002911 KMY92688.1 L27653 AE013599 AF145673 BT001354 BT099630 ACU64819.1
AKE50539.1 KQ460207 KPJ16759.1 NWSH01000300 PCG77630.1 AGBW02008321 OWR53675.1 GAIX01013897 JAA78663.1 GGFJ01002260 MBW51401.1 GGFK01001895 MBW35216.1 GGFK01001916 MBW35237.1 ADMH02000160 ETN67536.1 GGFJ01002259 MBW51400.1 APCN01000218 AAAB01008807 GGFJ01002261 MBW51402.1 GFDL01006186 JAV28859.1 GGFM01002081 MBW22832.1 GGFK01001902 MBW35223.1 ATLV01012372 KE524785 KFB36654.1 JXUM01042935 JXUM01042936 JXUM01042937 GAPW01000248 KQ561343 JAC13350.1 KXJ78899.1 AXCM01004236 GEDC01022015 JAS15283.1 AXCN02000593 GEDC01020516 GEDC01012156 JAS16782.1 JAS25142.1 GEDC01014581 GEDC01003943 JAS22717.1 JAS33355.1 EAL41738.3 GFDL01006222 JAV28823.1 GFDL01006232 JAV28813.1 GANO01001572 JAB58299.1 ABLF02041209 ABLF02041211 GFDF01001372 JAV12712.1 CH477410 EAT41468.1 GDAI01001796 JAI15807.1 UFQT01000063 SSX19272.1 DS232014 EDS31890.1 GFXV01007518 MBW19323.1 GAMC01016889 JAB89666.1 GAKP01021821 JAC37131.1 KK852854 KDR15026.1 GDHF01013540 JAI38774.1 NEVH01013227 PNF29623.1 KX147660 ARI45064.1 FX983726 BAX07225.1 GBXI01011096 GBXI01008564 JAD03196.1 JAD05728.1 DS235886 EEB20413.1 GALX01003377 JAB65089.1 GDHF01033073 GDHF01019749 JAI19241.1 JAI32565.1 GAKP01021822 JAC37130.1 GBXI01008380 JAD05912.1 KA646210 AFP60839.1 JXJN01007398 JXJN01007399 CH933808 EDW09504.2 CM002911 KMY92688.1 L27653 AE013599 AF145673 BT001354 BT099630 ACU64819.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000069272
+ More
UP000000673 UP000075882 UP000076407 UP000075903 UP000075840 UP000076408 UP000075885 UP000075884 UP000030765 UP000075900 UP000069940 UP000249989 UP000075883 UP000075886 UP000075920 UP000007062 UP000075880 UP000007819 UP000008820 UP000002320 UP000027135 UP000235965 UP000009046 UP000095301 UP000095300 UP000092460 UP000091820 UP000078200 UP000092445 UP000009192 UP000092443 UP000000803
UP000000673 UP000075882 UP000076407 UP000075903 UP000075840 UP000076408 UP000075885 UP000075884 UP000030765 UP000075900 UP000069940 UP000249989 UP000075883 UP000075886 UP000075920 UP000007062 UP000075880 UP000007819 UP000008820 UP000002320 UP000027135 UP000235965 UP000009046 UP000095301 UP000095300 UP000092460 UP000091820 UP000078200 UP000092445 UP000009192 UP000092443 UP000000803
Pfam
PF00365 PFK
Interpro
SUPFAM
SSF53784
SSF53784
ProteinModelPortal
H9J2I0
A0A2H1W9K4
A0A194PQ53
A0A0F6TNF4
A0A194RH42
A0A2A4K1T8
+ More
A0A212FIX8 S4NSV3 A0A2M4BEL6 A0A182FIA4 A0A2M4A324 A0A2M4A375 W5JTT0 A0A1C7CZ10 A0A2M4BEC6 A0A1Y9IXF3 A0A1Y9IRG4 A0A182I3Q9 A0A1Y9I2A6 A0A2M4BED7 A0A182XZI2 A0A1Q3FML9 A0A2M3Z2T4 A0A2M4A337 A0A240PN31 A0A1Y9H0Y5 A0A2K6V9F9 A0A084VFB0 A0A2K6V9F7 A0A182RFP8 A0A023EV74 A0A182MUK8 A0A1B6CPV7 A0A1Y9H9G2 A0A1B6DHL7 A0A182WIS9 A0A1B6DAI8 A0A1Y9IXX5 A0A1Y9IRZ8 A0A1S4GVQ5 A0A1Y9GIZ8 A0A182KRV4 A0A1Y9IS69 A0A1Y9GJI9 A0A182X6F6 Q5TX32 A0A1Q3FMX3 A0A240PNR8 A0A240PPG1 A0A1Q3FMW2 U5EUA4 A0A182P3I4 J9JNX0 A0A1Y9H0J2 A0A1Y9HAX6 A0A182NHK2 A0A1L8E2A1 A0A1Y9HAX5 A0A2C9GUF9 A0A1Y9HA64 A0A1Y9HAY6 Q174J0 A0A2C9GUI7 A0A1Y9IUN1 A0A1Y9IUM9 A0A0K8TN59 A0A182J6F9 A0A240PMF4 A0A336LMI2 B0WNP9 A0A2H8TYE0 W8B915 A0A034V1J7 A0A067QZR7 A0A0K8VIR0 A0A2J7QM39 A0A1W5YLK2 A0A2Z5RE15 A0A0A1X4W6 E0W457 A0A1I8MFB5 V5GLY8 A0A1I8MFB9 A0A1I8MFC8 A0A0K8V1T2 A0A034V5U4 A0A0A1X492 T1PDK0 A0A1I8NRU4 A0A1I8NRU1 A0A1I8NRU9 A0A1B0B232 A0A1A9WMU0 A0A1A9VY37 A0A1B0AGU6 B4KSJ9 A0A1A9YNY9 A0A0J9R9B9 P52034-3 C7LA80
A0A212FIX8 S4NSV3 A0A2M4BEL6 A0A182FIA4 A0A2M4A324 A0A2M4A375 W5JTT0 A0A1C7CZ10 A0A2M4BEC6 A0A1Y9IXF3 A0A1Y9IRG4 A0A182I3Q9 A0A1Y9I2A6 A0A2M4BED7 A0A182XZI2 A0A1Q3FML9 A0A2M3Z2T4 A0A2M4A337 A0A240PN31 A0A1Y9H0Y5 A0A2K6V9F9 A0A084VFB0 A0A2K6V9F7 A0A182RFP8 A0A023EV74 A0A182MUK8 A0A1B6CPV7 A0A1Y9H9G2 A0A1B6DHL7 A0A182WIS9 A0A1B6DAI8 A0A1Y9IXX5 A0A1Y9IRZ8 A0A1S4GVQ5 A0A1Y9GIZ8 A0A182KRV4 A0A1Y9IS69 A0A1Y9GJI9 A0A182X6F6 Q5TX32 A0A1Q3FMX3 A0A240PNR8 A0A240PPG1 A0A1Q3FMW2 U5EUA4 A0A182P3I4 J9JNX0 A0A1Y9H0J2 A0A1Y9HAX6 A0A182NHK2 A0A1L8E2A1 A0A1Y9HAX5 A0A2C9GUF9 A0A1Y9HA64 A0A1Y9HAY6 Q174J0 A0A2C9GUI7 A0A1Y9IUN1 A0A1Y9IUM9 A0A0K8TN59 A0A182J6F9 A0A240PMF4 A0A336LMI2 B0WNP9 A0A2H8TYE0 W8B915 A0A034V1J7 A0A067QZR7 A0A0K8VIR0 A0A2J7QM39 A0A1W5YLK2 A0A2Z5RE15 A0A0A1X4W6 E0W457 A0A1I8MFB5 V5GLY8 A0A1I8MFB9 A0A1I8MFC8 A0A0K8V1T2 A0A034V5U4 A0A0A1X492 T1PDK0 A0A1I8NRU4 A0A1I8NRU1 A0A1I8NRU9 A0A1B0B232 A0A1A9WMU0 A0A1A9VY37 A0A1B0AGU6 B4KSJ9 A0A1A9YNY9 A0A0J9R9B9 P52034-3 C7LA80
PDB
4OMT
E-value=0,
Score=2463
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
03018 RNA degradation - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
03018 RNA degradation - Bombyx mori (domestic silkworm)
GO
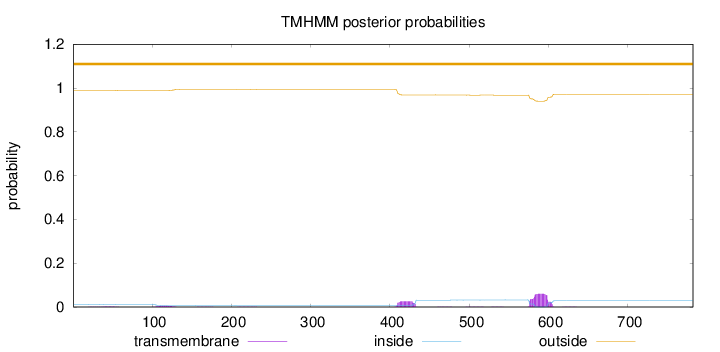
Topology
Subcellular location
Cytoplasm
Length:
782
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.08559
Exp number, first 60 AAs:
0.0302
Total prob of N-in:
0.01212
outside
1 - 782
Population Genetic Test Statistics
Pi
305.224967
Theta
194.923481
Tajima's D
1.609517
CLR
0.00316
CSRT
0.809459527023649
Interpretation
Uncertain
