Gene
KWMTBOMO02742
Annotation
PREDICTED:_uncharacterized_protein_LOC106135477_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.905
Sequence
CDS
ATGCTGGACGAGAACAGTGGAAAATCAGTTTGTCGTATTCCAATTGATAGAGATGATCCTACTGGGTGCGGTGGACTCTGTACAATGGAATTAGAAGCTTGTCATTTGGTAGACAAGGAATTAGGTGTTCGTGTTTGTAGACTTTTAACACTAGCAACGTGTTCACCGGAAGAATGGCGTTGTCGTAATGGTTTATGTGTTCCTACAGAAAGCAGATGCAATGGTTCAGTAGAATGCTATGATCGATCAGACGAAGTACATTGCGACTGTGATCTTACAAAGCAGTTTCGCTGCGGGAATTTAATGTCTTGTTTTTCTAACATGAAGCTCTGCGATGGGATCGTCGACTGTTGGGATGGGTTCGACGAACGTAATTGCACTGAATGTCCGCAAAACCAATCACCGTGTAAAGATGGCCAATGTTTTTTATCATCAAGATTTTGTGATGGCTTCGTAGATTGCGAAGATCGCAGTGATGAACCAGATGGCTGTGGAGGATCGTGTAGGAAGCACGAACTGCGTTGCTTGAATCAACGTTGCGTACCTCTCACAGTAAAATGCGACGGTCAAGATAACTGCGGGGATGCTTCCGATGAATTAAATTGCTTTATGACCTAA
Protein
MLDENSGKSVCRIPIDRDDPTGCGGLCTMELEACHLVDKELGVRVCRLLTLATCSPEEWRCRNGLCVPTESRCNGSVECYDRSDEVHCDCDLTKQFRCGNLMSCFSNMKLCDGIVDCWDGFDERNCTECPQNQSPCKDGQCFLSSRFCDGFVDCEDRSDEPDGCGGSCRKHELRCLNQRCVPLTVKCDGQDNCGDASDELNCFMT
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A0A2A4JFH0
A0A2H1WWB0
A0A212FIX6
A0A194PNG6
A0A194RGN8
A0A0L7KQG3
+ More
A0A067R6L8 A0A2R7W8F0 A0A1V1FIX2 A0A1B6E7T3 A0A1B6IRG4 A0A1B6FME6 A0A0C9RJL0 A0A087ZWG5 E9J535 A0A0C9RMW1 A0A154PT49 A0A0M9A8B8 A0A0L7QVF9 A0A151X4U8 A0A131XLH1 A0A2R5L910 E2A7C9 A0A195D9E2 A0A195BPT4 A0A158NE10 A0A151IM54 A0A1E1X781 A0A195ESG9 A0A1E1XKP3 F4X1S2 A0A131YWU6 B7QKS7 K7JL11 T1IV70 A0A026W8D1 X1X7S3 J9K9N2 A0A2S2NZ55 A0A1Y1NCU5 A0A1V9XKA2 N6URR0 A0A1B6LW04 A0A2P8Y982 T1F504 A0A1B6F0K1 A0A1B6HQR8 A0A1B6K3W3 A0A0L8GZ95 A0A1W4WI19 A0A1B6CE74 A0A1S4ENM7 A0A1J1ISF7 A0A2J7PDY1 A0A0V1L9H1 A0A0V1L9P5 A0A0V0TQN3 A0A0V1P7W8 A0A0V0Y9I0 A0A0V0Y973 A0A0V1L9Q7 A0A0V1A039 A0A0V0S1Z7 A0A0V0TQK3 A0A0V1LAG3 A0A0V1H9H1 A0A0V0TQR2 A0A0V0YA48 A0A0V0S2A3 A0A0V0VJJ6 A0A0V1D5C2 A0A0V1D5B8 A0A1Y3ER74 A0A0V1D557 A0A0V0S2D0 A0A0V1D6H8 A0A0V1JSM3 A0A0V0Y9W0 A0A0V0TRW7 A0A0M3QYX2 A0A0P5C164 A0A2I4BNN1 A0A0P5CBY8 W5JU70 A0A182FBA2 A0A194Q149 A0A3B5QXR6 A7RXB7 A0A0V1L9K7 A0A023FEK7 A0A0V0S2W6 A0A0N1ID44 A0A1E1WWY3 A0A2C9LNM5 A0A2G8KY87 A0A3B5QWI6 A0A2P2HZD4 A0A3P7L8R5
A0A067R6L8 A0A2R7W8F0 A0A1V1FIX2 A0A1B6E7T3 A0A1B6IRG4 A0A1B6FME6 A0A0C9RJL0 A0A087ZWG5 E9J535 A0A0C9RMW1 A0A154PT49 A0A0M9A8B8 A0A0L7QVF9 A0A151X4U8 A0A131XLH1 A0A2R5L910 E2A7C9 A0A195D9E2 A0A195BPT4 A0A158NE10 A0A151IM54 A0A1E1X781 A0A195ESG9 A0A1E1XKP3 F4X1S2 A0A131YWU6 B7QKS7 K7JL11 T1IV70 A0A026W8D1 X1X7S3 J9K9N2 A0A2S2NZ55 A0A1Y1NCU5 A0A1V9XKA2 N6URR0 A0A1B6LW04 A0A2P8Y982 T1F504 A0A1B6F0K1 A0A1B6HQR8 A0A1B6K3W3 A0A0L8GZ95 A0A1W4WI19 A0A1B6CE74 A0A1S4ENM7 A0A1J1ISF7 A0A2J7PDY1 A0A0V1L9H1 A0A0V1L9P5 A0A0V0TQN3 A0A0V1P7W8 A0A0V0Y9I0 A0A0V0Y973 A0A0V1L9Q7 A0A0V1A039 A0A0V0S1Z7 A0A0V0TQK3 A0A0V1LAG3 A0A0V1H9H1 A0A0V0TQR2 A0A0V0YA48 A0A0V0S2A3 A0A0V0VJJ6 A0A0V1D5C2 A0A0V1D5B8 A0A1Y3ER74 A0A0V1D557 A0A0V0S2D0 A0A0V1D6H8 A0A0V1JSM3 A0A0V0Y9W0 A0A0V0TRW7 A0A0M3QYX2 A0A0P5C164 A0A2I4BNN1 A0A0P5CBY8 W5JU70 A0A182FBA2 A0A194Q149 A0A3B5QXR6 A7RXB7 A0A0V1L9K7 A0A023FEK7 A0A0V0S2W6 A0A0N1ID44 A0A1E1WWY3 A0A2C9LNM5 A0A2G8KY87 A0A3B5QWI6 A0A2P2HZD4 A0A3P7L8R5
Pubmed
EMBL
NWSH01001776
PCG70180.1
ODYU01011505
SOQ57277.1
AGBW02008321
OWR53681.1
+ More
KQ459597 KPI94862.1 KQ460207 KPJ16752.1 JTDY01007446 KOB65194.1 KK852854 KDR15023.1 KK854458 PTY16007.1 FX985388 BAX07401.1 GEDC01003297 JAS34001.1 GECU01018213 JAS89493.1 GECZ01028103 GECZ01018420 JAS41666.1 JAS51349.1 GBYB01008385 JAG78152.1 GL768123 EFZ12061.1 GBYB01008386 JAG78153.1 KQ435078 KZC14438.1 KQ435713 KOX79305.1 KQ414727 KOC62544.1 KQ982548 KYQ55324.1 GEFH01001184 JAP67397.1 GGLE01001833 MBY05959.1 GL437329 EFN70626.1 KQ981153 KYN09019.1 KQ976424 KYM88520.1 ADTU01013089 ADTU01013090 KQ977074 KYN05944.1 GFAC01004120 JAT95068.1 KQ981993 KYN30834.1 GFAA01003532 JAT99902.1 GL888551 EGI59579.1 GEDV01005582 JAP82975.1 ABJB010037934 DS961818 EEC19449.1 JH431575 KK107347 EZA52223.1 ABLF02024464 ABLF02010464 GGMR01009874 MBY22493.1 GEZM01010420 JAV94056.1 MNPL01009286 OQR73788.1 APGK01019082 KB740092 KB632429 ENN81422.1 ERL95472.1 GEBQ01012140 JAT27837.1 PYGN01000786 PSN40826.1 AMQM01004032 KB096411 ESO05065.1 GECZ01026042 GECZ01000340 JAS43727.1 JAS69429.1 GECU01030703 GECU01017883 JAS77003.1 JAS89823.1 GECU01001554 JAT06153.1 KQ419813 KOF82288.1 GEDC01025559 JAS11739.1 CVRI01000059 CRL03072.1 NEVH01026122 PNF14536.1 JYDW01000098 KRZ56207.1 KRZ56211.1 JYDJ01000176 KRX41248.1 JYDM01000035 KRZ92327.1 JYDU01000036 KRX97041.1 KRX97044.1 KRZ56209.1 JYDQ01000052 KRY18061.1 JYDL01000045 KRX20790.1 KRX41250.1 KRZ56210.1 JYDP01000102 KRZ07432.1 KRX41249.1 KRX97045.1 KRX20788.1 JYDN01000029 KRX63675.1 JYDI01000041 KRY56582.1 KRY56580.1 LVZM01003959 OUC47644.1 KRY56583.1 KRX20791.1 KRY56579.1 JYDV01000052 KRZ37912.1 KRX97043.1 KRX41251.1 CP012528 ALC48423.1 GDIP01177611 JAJ45791.1 GDIP01177610 JAJ45792.1 ADMH02000120 ETN67681.1 KQ459590 KPI97125.1 DS469549 EDO43937.1 KRZ56208.1 GBBK01005163 JAC19319.1 KRX20789.1 KQ459692 KPJ20448.1 GFAC01007892 JAT91296.1 MRZV01000307 PIK52967.1 IACF01001438 LAB67138.1 UYRU01057118 VDN13705.1
KQ459597 KPI94862.1 KQ460207 KPJ16752.1 JTDY01007446 KOB65194.1 KK852854 KDR15023.1 KK854458 PTY16007.1 FX985388 BAX07401.1 GEDC01003297 JAS34001.1 GECU01018213 JAS89493.1 GECZ01028103 GECZ01018420 JAS41666.1 JAS51349.1 GBYB01008385 JAG78152.1 GL768123 EFZ12061.1 GBYB01008386 JAG78153.1 KQ435078 KZC14438.1 KQ435713 KOX79305.1 KQ414727 KOC62544.1 KQ982548 KYQ55324.1 GEFH01001184 JAP67397.1 GGLE01001833 MBY05959.1 GL437329 EFN70626.1 KQ981153 KYN09019.1 KQ976424 KYM88520.1 ADTU01013089 ADTU01013090 KQ977074 KYN05944.1 GFAC01004120 JAT95068.1 KQ981993 KYN30834.1 GFAA01003532 JAT99902.1 GL888551 EGI59579.1 GEDV01005582 JAP82975.1 ABJB010037934 DS961818 EEC19449.1 JH431575 KK107347 EZA52223.1 ABLF02024464 ABLF02010464 GGMR01009874 MBY22493.1 GEZM01010420 JAV94056.1 MNPL01009286 OQR73788.1 APGK01019082 KB740092 KB632429 ENN81422.1 ERL95472.1 GEBQ01012140 JAT27837.1 PYGN01000786 PSN40826.1 AMQM01004032 KB096411 ESO05065.1 GECZ01026042 GECZ01000340 JAS43727.1 JAS69429.1 GECU01030703 GECU01017883 JAS77003.1 JAS89823.1 GECU01001554 JAT06153.1 KQ419813 KOF82288.1 GEDC01025559 JAS11739.1 CVRI01000059 CRL03072.1 NEVH01026122 PNF14536.1 JYDW01000098 KRZ56207.1 KRZ56211.1 JYDJ01000176 KRX41248.1 JYDM01000035 KRZ92327.1 JYDU01000036 KRX97041.1 KRX97044.1 KRZ56209.1 JYDQ01000052 KRY18061.1 JYDL01000045 KRX20790.1 KRX41250.1 KRZ56210.1 JYDP01000102 KRZ07432.1 KRX41249.1 KRX97045.1 KRX20788.1 JYDN01000029 KRX63675.1 JYDI01000041 KRY56582.1 KRY56580.1 LVZM01003959 OUC47644.1 KRY56583.1 KRX20791.1 KRY56579.1 JYDV01000052 KRZ37912.1 KRX97043.1 KRX41251.1 CP012528 ALC48423.1 GDIP01177611 JAJ45791.1 GDIP01177610 JAJ45792.1 ADMH02000120 ETN67681.1 KQ459590 KPI97125.1 DS469549 EDO43937.1 KRZ56208.1 GBBK01005163 JAC19319.1 KRX20789.1 KQ459692 KPJ20448.1 GFAC01007892 JAT91296.1 MRZV01000307 PIK52967.1 IACF01001438 LAB67138.1 UYRU01057118 VDN13705.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
UP000027135
+ More
UP000005203 UP000076502 UP000053105 UP000053825 UP000075809 UP000000311 UP000078492 UP000078540 UP000005205 UP000078542 UP000078541 UP000007755 UP000001555 UP000002358 UP000053097 UP000007819 UP000192247 UP000019118 UP000030742 UP000245037 UP000015101 UP000053454 UP000192223 UP000079169 UP000183832 UP000235965 UP000054721 UP000055048 UP000054924 UP000054815 UP000054783 UP000054630 UP000055024 UP000054681 UP000054653 UP000243006 UP000054826 UP000092553 UP000192220 UP000000673 UP000069272 UP000002852 UP000001593 UP000076420 UP000230750
UP000005203 UP000076502 UP000053105 UP000053825 UP000075809 UP000000311 UP000078492 UP000078540 UP000005205 UP000078542 UP000078541 UP000007755 UP000001555 UP000002358 UP000053097 UP000007819 UP000192247 UP000019118 UP000030742 UP000245037 UP000015101 UP000053454 UP000192223 UP000079169 UP000183832 UP000235965 UP000054721 UP000055048 UP000054924 UP000054815 UP000054783 UP000054630 UP000055024 UP000054681 UP000054653 UP000243006 UP000054826 UP000092553 UP000192220 UP000000673 UP000069272 UP000002852 UP000001593 UP000076420 UP000230750
Pfam
PF00057 Ldl_recept_a
+ More
PF01390 SEA
PF07974 EGF_2
PF00008 EGF
PF00058 Ldl_recept_b
PF07679 I-set
PF00053 Laminin_EGF
PF00052 Laminin_B
PF02931 Neur_chan_LBD
PF02932 Neur_chan_memb
PF16472 DUF5050
PF12662 cEGF
PF07645 EGF_CA
PF00054 Laminin_G_1
PF02210 Laminin_G_2
PF00047 ig
PF13895 Ig_2
PF00092 VWA
PF00530 SRCR
PF01390 SEA
PF07974 EGF_2
PF00008 EGF
PF00058 Ldl_recept_b
PF07679 I-set
PF00053 Laminin_EGF
PF00052 Laminin_B
PF02931 Neur_chan_LBD
PF02932 Neur_chan_memb
PF16472 DUF5050
PF12662 cEGF
PF07645 EGF_CA
PF00054 Laminin_G_1
PF02210 Laminin_G_2
PF00047 ig
PF13895 Ig_2
PF00092 VWA
PF00530 SRCR
Interpro
IPR036055
LDL_receptor-like_sf
+ More
IPR002172 LDrepeatLR_classA_rpt
IPR023415 LDLR_class-A_CS
IPR036383 TSP1_rpt_sf
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
IPR040093 LRAD1
IPR000884 TSP1_rpt
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR009030 Growth_fac_rcpt_cys_sf
IPR013098 Ig_I-set
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR003598 Ig_sub2
IPR036719 Neuro-gated_channel_TM_sf
IPR032485 DUF5050
IPR036734 Neur_chan_lig-bd_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR006201 Neur_channel
IPR026823 cEGF
IPR018000 Neurotransmitter_ion_chnl_CS
IPR013106 Ig_V-set
IPR001791 Laminin_G
IPR013151 Immunoglobulin
IPR013320 ConA-like_dom_sf
IPR002035 VWF_A
IPR036465 vWFA_dom_sf
IPR017448 SRCR-like_dom
IPR001190 SRCR
IPR036772 SRCR-like_dom_sf
IPR002172 LDrepeatLR_classA_rpt
IPR023415 LDLR_class-A_CS
IPR036383 TSP1_rpt_sf
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
IPR040093 LRAD1
IPR000884 TSP1_rpt
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR009030 Growth_fac_rcpt_cys_sf
IPR013098 Ig_I-set
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR003598 Ig_sub2
IPR036719 Neuro-gated_channel_TM_sf
IPR032485 DUF5050
IPR036734 Neur_chan_lig-bd_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR006201 Neur_channel
IPR026823 cEGF
IPR018000 Neurotransmitter_ion_chnl_CS
IPR013106 Ig_V-set
IPR001791 Laminin_G
IPR013151 Immunoglobulin
IPR013320 ConA-like_dom_sf
IPR002035 VWF_A
IPR036465 vWFA_dom_sf
IPR017448 SRCR-like_dom
IPR001190 SRCR
IPR036772 SRCR-like_dom_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2A4JFH0
A0A2H1WWB0
A0A212FIX6
A0A194PNG6
A0A194RGN8
A0A0L7KQG3
+ More
A0A067R6L8 A0A2R7W8F0 A0A1V1FIX2 A0A1B6E7T3 A0A1B6IRG4 A0A1B6FME6 A0A0C9RJL0 A0A087ZWG5 E9J535 A0A0C9RMW1 A0A154PT49 A0A0M9A8B8 A0A0L7QVF9 A0A151X4U8 A0A131XLH1 A0A2R5L910 E2A7C9 A0A195D9E2 A0A195BPT4 A0A158NE10 A0A151IM54 A0A1E1X781 A0A195ESG9 A0A1E1XKP3 F4X1S2 A0A131YWU6 B7QKS7 K7JL11 T1IV70 A0A026W8D1 X1X7S3 J9K9N2 A0A2S2NZ55 A0A1Y1NCU5 A0A1V9XKA2 N6URR0 A0A1B6LW04 A0A2P8Y982 T1F504 A0A1B6F0K1 A0A1B6HQR8 A0A1B6K3W3 A0A0L8GZ95 A0A1W4WI19 A0A1B6CE74 A0A1S4ENM7 A0A1J1ISF7 A0A2J7PDY1 A0A0V1L9H1 A0A0V1L9P5 A0A0V0TQN3 A0A0V1P7W8 A0A0V0Y9I0 A0A0V0Y973 A0A0V1L9Q7 A0A0V1A039 A0A0V0S1Z7 A0A0V0TQK3 A0A0V1LAG3 A0A0V1H9H1 A0A0V0TQR2 A0A0V0YA48 A0A0V0S2A3 A0A0V0VJJ6 A0A0V1D5C2 A0A0V1D5B8 A0A1Y3ER74 A0A0V1D557 A0A0V0S2D0 A0A0V1D6H8 A0A0V1JSM3 A0A0V0Y9W0 A0A0V0TRW7 A0A0M3QYX2 A0A0P5C164 A0A2I4BNN1 A0A0P5CBY8 W5JU70 A0A182FBA2 A0A194Q149 A0A3B5QXR6 A7RXB7 A0A0V1L9K7 A0A023FEK7 A0A0V0S2W6 A0A0N1ID44 A0A1E1WWY3 A0A2C9LNM5 A0A2G8KY87 A0A3B5QWI6 A0A2P2HZD4 A0A3P7L8R5
A0A067R6L8 A0A2R7W8F0 A0A1V1FIX2 A0A1B6E7T3 A0A1B6IRG4 A0A1B6FME6 A0A0C9RJL0 A0A087ZWG5 E9J535 A0A0C9RMW1 A0A154PT49 A0A0M9A8B8 A0A0L7QVF9 A0A151X4U8 A0A131XLH1 A0A2R5L910 E2A7C9 A0A195D9E2 A0A195BPT4 A0A158NE10 A0A151IM54 A0A1E1X781 A0A195ESG9 A0A1E1XKP3 F4X1S2 A0A131YWU6 B7QKS7 K7JL11 T1IV70 A0A026W8D1 X1X7S3 J9K9N2 A0A2S2NZ55 A0A1Y1NCU5 A0A1V9XKA2 N6URR0 A0A1B6LW04 A0A2P8Y982 T1F504 A0A1B6F0K1 A0A1B6HQR8 A0A1B6K3W3 A0A0L8GZ95 A0A1W4WI19 A0A1B6CE74 A0A1S4ENM7 A0A1J1ISF7 A0A2J7PDY1 A0A0V1L9H1 A0A0V1L9P5 A0A0V0TQN3 A0A0V1P7W8 A0A0V0Y9I0 A0A0V0Y973 A0A0V1L9Q7 A0A0V1A039 A0A0V0S1Z7 A0A0V0TQK3 A0A0V1LAG3 A0A0V1H9H1 A0A0V0TQR2 A0A0V0YA48 A0A0V0S2A3 A0A0V0VJJ6 A0A0V1D5C2 A0A0V1D5B8 A0A1Y3ER74 A0A0V1D557 A0A0V0S2D0 A0A0V1D6H8 A0A0V1JSM3 A0A0V0Y9W0 A0A0V0TRW7 A0A0M3QYX2 A0A0P5C164 A0A2I4BNN1 A0A0P5CBY8 W5JU70 A0A182FBA2 A0A194Q149 A0A3B5QXR6 A7RXB7 A0A0V1L9K7 A0A023FEK7 A0A0V0S2W6 A0A0N1ID44 A0A1E1WWY3 A0A2C9LNM5 A0A2G8KY87 A0A3B5QWI6 A0A2P2HZD4 A0A3P7L8R5
PDB
3M0C
E-value=5.63913e-16,
Score=202
Ontologies
KEGG
GO
PANTHER
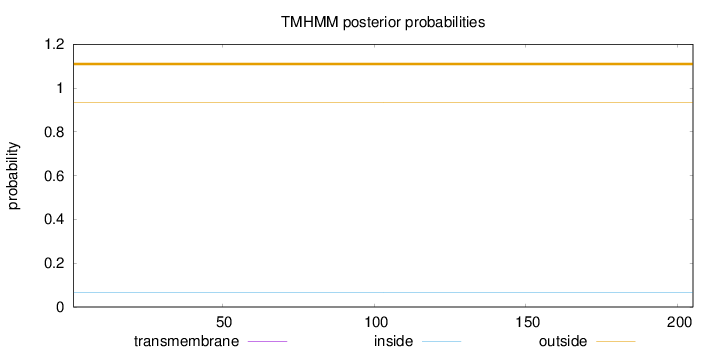
Topology
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06704
outside
1 - 205
Population Genetic Test Statistics
Pi
228.003365
Theta
175.529603
Tajima's D
1.22646
CLR
0
CSRT
0.72246387680616
Interpretation
Uncertain
