Gene
KWMTBOMO02741
Pre Gene Modal
BGIBMGA003720
Annotation
PREDICTED:_sphingomyelin_phosphodiesterase_isoform_X1_[Bombyx_mori]
Full name
Sphingomyelin phosphodiesterase
Location in the cell
Lysosomal Reliability : 1.368
Sequence
CDS
ATGATCATTAGAATATTATGGCTTCTACTACTGGTGATTGTTTCAGTAAGCCCCTGGACATTAGGTAACCCAATATTCCTACAGAAGCCTGGTAAGCCAAACAGACCTGTGATAAAATGGAATCCTCCAACGGACACGTGGAACTGGACTACGAATCCCCGAAACGAGGAACCGTTTTTTGTACCGTCAAATTTTACGCATAAATTGCCACCATCGGCCAATGAAACCGAACTCATAGCTCACATGGCAGCTTTGAAACGTAACTATTCGCGGCTCCTCTGGGATAACGATAAAGGCACGAATTTACCTATGTTCGTTGAAAAAGCATTAAAACTACTCAACTTGAAACAGGTTTATTACGAAGTTGAGCATTCGGTGATGTCCAAAGTGTCGTGCACCGCGTGTAAGGCAGGCGCAGGACTGCTGCAGCACTATATAAGGCTCGGTAAAAGTAAAGATGAGATCAACAAAATGATTTTTCAATTTTGCGTTTCTTTAAAGATTCAATCCGCTCGAGTTTGTGAAGGAATAACGAGATTATTCGGGAGCGAAGTGGTATATGTTTTGAAAAGAATAACAATTGGCCCGGACGAAATATGCAGTTTTGTGATTGGTGACGCTTGTGGCGATGTTTATAATCCATATCACGAATGGGAAGTGGCTTTCCCTCCGGTGCCCAAGCCGACTGTGCGACCTCTCGAATCACCGTTAGAAAAGGCACCCACATTCAAAGTATTGCAGATTTCAGATACGCATTTCGATCCTTATTATGCAGAAGGTGCTAATGCCGAGTGTAATGAACCACTTTGTTGTCGTGCGTCTAACGGACCCGCTCTGACCCCGGGTGGCGGAGCAGGACGTTGGGGAGACTATAGAAAATGTGACACACCGAAGCGCACTATTGATCACATGTTGAAGCACATTGCTGACACACATACAGACATAGACTATATATTGTGGACCGGAGATTTACCACCACATGACGTTTGGAATCAAACGAAGGAAGAAAATCTCAAAGTTTTACAAGAGACTGTTGCACAAATGTCAGATATGTTTCCTGGAGTACCAATTTTCCCAGCTCTAGGAAATCACGAATCTTCTCCGGTAAATAGTTTTCCACCGCCCTACATATCTTCGCCGGACCAAAACATCGCTTGGCTATATGACGAACTTGACACGCAATGGCGACGTTGGCTTCCTGCTGGCGTGTCGCACACCGTACGCCGGGGCGCATTCTACTCGGTACTAGTGAGGCCTGGCTTCCGTATAATATCGCTTAACATGAACTACTGCAATAATAAAAACTGGTTAGTTTTTGTAATTATTACCCAAAAAATTTTTTTTACGTTTTAA
Protein
MIIRILWLLLLVIVSVSPWTLGNPIFLQKPGKPNRPVIKWNPPTDTWNWTTNPRNEEPFFVPSNFTHKLPPSANETELIAHMAALKRNYSRLLWDNDKGTNLPMFVEKALKLLNLKQVYYEVEHSVMSKVSCTACKAGAGLLQHYIRLGKSKDEINKMIFQFCVSLKIQSARVCEGITRLFGSEVVYVLKRITIGPDEICSFVIGDACGDVYNPYHEWEVAFPPVPKPTVRPLESPLEKAPTFKVLQISDTHFDPYYAEGANAECNEPLCCRASNGPALTPGGGAGRWGDYRKCDTPKRTIDHMLKHIADTHTDIDYILWTGDLPPHDVWNQTKEENLKVLQETVAQMSDMFPGVPIFPALGNHESSPVNSFPPPYISSPDQNIAWLYDELDTQWRRWLPAGVSHTVRRGAFYSVLVRPGFRIISLNMNYCNNKNWLVFVIITQKIFFTF
Summary
Description
Converts sphingomyelin to ceramide.
Catalytic Activity
H2O + sphingomyelin = an N-acylsphing-4-enine + H(+) + phosphocholine
Cofactor
Zn(2+)
Similarity
Belongs to the acid sphingomyelinase family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 1 family.
Feature
chain Sphingomyelin phosphodiesterase
Uniprot
A0A2H1WW48
A0A194RHF1
A0A194PN53
I4DM60
A0A212FIX9
A0A182U186
+ More
A0A182XJQ6 A0A182PMW3 A0A182VGX6 A0A182L586 A0A182IIK0 A0A182Q6E2 A0A182IIK1 Q7PZJ1 A0A182Y9I9 A0A182KEX5 A0A182M166 A0A084WIG8 A0A182RXV4 A0A182JHX6 A0A182N2R8 A0A182WQP9 A0A182SBY3 A0A182RXV5 A0A182WQP8 A0A1L8DPQ0 A0A1L8DQ04 A0A1L8DPS5 A0A1S4F4J5 Q17FG9 W5JWZ2 A0A182FZW9 Q17FG8 A0A182FZW8 Q17FH0 W8BSV3 W8ATX5 A0A2M4ALK1 A0A2M4ANM1 A0A2M4BFI8 W8BG77 A0A2M4ALB3 A0A2M4ALQ8 A0A2M4BGF7 A0A2K6VB97 A0A034V391 A0A0K8UKX6 D6WVW3 A0A139WDF2 A0A0A1XFR8 A0A0A1XCZ7 A0A0K8VNE0 A0A034V153 A0A0K8U320 B0WS01 A0A0T6B856 A0A1A9ZW88 A0A1B0FF57 U5TZ69 A0A1Q3FC40 A0A1Q3FEJ9 A0A1Q3FEE2 A0A1Q3FD90 A0A1Q3FBV2 A0A1I8P9V5 A0A1I8P9N2 A0A1I8P9M7 D3TNH1 A0A1A9UKZ8 A0A1A9X4A2 A0A1B0BYE8 A0A1W4UZV4 A0A1A9YGB8 A0A1W4V1F0 A0A0J9RJW2 A0A0J9UA65 A0A0J9RKB0 A0A0J9RKG8 B4QBX0 B3NQH4 A0A336MAU4 A0A026WT08 A0A336MIZ9 A0A1B0GGU0 B4PAW9 B4H694 A0A0R1DTP2 B4MYD9 Q28WU3 N6VFZ6 A0A151WVL7 B4J4T2 A0A3B0IYW2 A0A3B0J2R2 E2C0J8 A0A3B0IZ62 J9JZU0 A0A0M4EDA7 A0A0Q9XK69 B4KMI1
A0A182XJQ6 A0A182PMW3 A0A182VGX6 A0A182L586 A0A182IIK0 A0A182Q6E2 A0A182IIK1 Q7PZJ1 A0A182Y9I9 A0A182KEX5 A0A182M166 A0A084WIG8 A0A182RXV4 A0A182JHX6 A0A182N2R8 A0A182WQP9 A0A182SBY3 A0A182RXV5 A0A182WQP8 A0A1L8DPQ0 A0A1L8DQ04 A0A1L8DPS5 A0A1S4F4J5 Q17FG9 W5JWZ2 A0A182FZW9 Q17FG8 A0A182FZW8 Q17FH0 W8BSV3 W8ATX5 A0A2M4ALK1 A0A2M4ANM1 A0A2M4BFI8 W8BG77 A0A2M4ALB3 A0A2M4ALQ8 A0A2M4BGF7 A0A2K6VB97 A0A034V391 A0A0K8UKX6 D6WVW3 A0A139WDF2 A0A0A1XFR8 A0A0A1XCZ7 A0A0K8VNE0 A0A034V153 A0A0K8U320 B0WS01 A0A0T6B856 A0A1A9ZW88 A0A1B0FF57 U5TZ69 A0A1Q3FC40 A0A1Q3FEJ9 A0A1Q3FEE2 A0A1Q3FD90 A0A1Q3FBV2 A0A1I8P9V5 A0A1I8P9N2 A0A1I8P9M7 D3TNH1 A0A1A9UKZ8 A0A1A9X4A2 A0A1B0BYE8 A0A1W4UZV4 A0A1A9YGB8 A0A1W4V1F0 A0A0J9RJW2 A0A0J9UA65 A0A0J9RKB0 A0A0J9RKG8 B4QBX0 B3NQH4 A0A336MAU4 A0A026WT08 A0A336MIZ9 A0A1B0GGU0 B4PAW9 B4H694 A0A0R1DTP2 B4MYD9 Q28WU3 N6VFZ6 A0A151WVL7 B4J4T2 A0A3B0IYW2 A0A3B0J2R2 E2C0J8 A0A3B0IZ62 J9JZU0 A0A0M4EDA7 A0A0Q9XK69 B4KMI1
EC Number
3.1.4.12
Pubmed
EMBL
ODYU01011505
SOQ57278.1
KQ460207
KPJ16750.1
KQ459597
KPI94861.1
+ More
AK402378 BAM19000.1 AGBW02008321 OWR53682.1 APCN01002429 AXCN02001087 AAAB01008986 EAA00261.3 AXCM01001841 ATLV01023931 KE525347 KFB50012.1 GFDF01005626 JAV08458.1 GFDF01005627 JAV08457.1 GFDF01005628 JAV08456.1 CH477271 EAT45275.1 ADMH02000120 ETN67704.1 EAT45276.1 EAT45277.1 GAMC01014261 JAB92294.1 GAMC01014260 JAB92295.1 GGFK01008291 MBW41612.1 GGFK01009011 MBW42332.1 GGFJ01002580 MBW51721.1 GAMC01014259 JAB92296.1 GGFK01008248 MBW41569.1 GGFK01008383 MBW41704.1 GGFJ01002747 MBW51888.1 GAKP01021948 JAC37004.1 GDHF01024977 JAI27337.1 KQ971358 EFA08230.2 KYB25998.1 GBXI01004959 JAD09333.1 GBXI01005491 JAD08801.1 GDHF01011917 JAI40397.1 GAKP01021946 JAC37006.1 GDHF01031381 JAI20933.1 DS232063 EDS33619.1 LJIG01009347 KRT83261.1 CCAG010006382 CCAG010006383 KF417890 AGZ15175.1 GFDL01009901 JAV25144.1 GFDL01009087 JAV25958.1 GFDL01009122 JAV25923.1 GFDL01009552 JAV25493.1 GFDL01010053 JAV24992.1 EZ422973 JQ308535 ADD19249.1 AFJ68090.1 JXJN01022601 CM002911 KMY96303.1 KMY96300.1 KMY96301.1 KMY96302.1 CM000362 EDX08525.1 CH954179 EDV56977.1 UFQT01000641 SSX26029.1 KK107119 QOIP01000009 EZA58816.1 RLU18737.1 SSX26028.1 AJWK01000745 AJWK01000746 AJWK01000747 CM000158 EDW92509.1 CH479213 EDW33318.1 KRK00555.1 CH963894 EDW77128.2 CM000071 EAL26573.2 ENO01975.2 KQ982699 KYQ51950.1 CH916367 EDW00628.1 OUUW01000001 SPP73394.1 SPP73393.1 GL451800 EFN78515.1 SPP73395.1 ABLF02039753 CP012524 ALC41877.1 CH933808 KRG05506.1 EDW10828.1
AK402378 BAM19000.1 AGBW02008321 OWR53682.1 APCN01002429 AXCN02001087 AAAB01008986 EAA00261.3 AXCM01001841 ATLV01023931 KE525347 KFB50012.1 GFDF01005626 JAV08458.1 GFDF01005627 JAV08457.1 GFDF01005628 JAV08456.1 CH477271 EAT45275.1 ADMH02000120 ETN67704.1 EAT45276.1 EAT45277.1 GAMC01014261 JAB92294.1 GAMC01014260 JAB92295.1 GGFK01008291 MBW41612.1 GGFK01009011 MBW42332.1 GGFJ01002580 MBW51721.1 GAMC01014259 JAB92296.1 GGFK01008248 MBW41569.1 GGFK01008383 MBW41704.1 GGFJ01002747 MBW51888.1 GAKP01021948 JAC37004.1 GDHF01024977 JAI27337.1 KQ971358 EFA08230.2 KYB25998.1 GBXI01004959 JAD09333.1 GBXI01005491 JAD08801.1 GDHF01011917 JAI40397.1 GAKP01021946 JAC37006.1 GDHF01031381 JAI20933.1 DS232063 EDS33619.1 LJIG01009347 KRT83261.1 CCAG010006382 CCAG010006383 KF417890 AGZ15175.1 GFDL01009901 JAV25144.1 GFDL01009087 JAV25958.1 GFDL01009122 JAV25923.1 GFDL01009552 JAV25493.1 GFDL01010053 JAV24992.1 EZ422973 JQ308535 ADD19249.1 AFJ68090.1 JXJN01022601 CM002911 KMY96303.1 KMY96300.1 KMY96301.1 KMY96302.1 CM000362 EDX08525.1 CH954179 EDV56977.1 UFQT01000641 SSX26029.1 KK107119 QOIP01000009 EZA58816.1 RLU18737.1 SSX26028.1 AJWK01000745 AJWK01000746 AJWK01000747 CM000158 EDW92509.1 CH479213 EDW33318.1 KRK00555.1 CH963894 EDW77128.2 CM000071 EAL26573.2 ENO01975.2 KQ982699 KYQ51950.1 CH916367 EDW00628.1 OUUW01000001 SPP73394.1 SPP73393.1 GL451800 EFN78515.1 SPP73395.1 ABLF02039753 CP012524 ALC41877.1 CH933808 KRG05506.1 EDW10828.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000075902
UP000076407
UP000075885
+ More
UP000075903 UP000075882 UP000075840 UP000075886 UP000007062 UP000076408 UP000075881 UP000075883 UP000030765 UP000075900 UP000075880 UP000075884 UP000075920 UP000075901 UP000008820 UP000000673 UP000069272 UP000007266 UP000002320 UP000092445 UP000092444 UP000095300 UP000078200 UP000091820 UP000092460 UP000192221 UP000092443 UP000000304 UP000008711 UP000053097 UP000279307 UP000092461 UP000002282 UP000008744 UP000007798 UP000001819 UP000075809 UP000001070 UP000268350 UP000008237 UP000007819 UP000092553 UP000009192
UP000075903 UP000075882 UP000075840 UP000075886 UP000007062 UP000076408 UP000075881 UP000075883 UP000030765 UP000075900 UP000075880 UP000075884 UP000075920 UP000075901 UP000008820 UP000000673 UP000069272 UP000007266 UP000002320 UP000092445 UP000092444 UP000095300 UP000078200 UP000091820 UP000092460 UP000192221 UP000092443 UP000000304 UP000008711 UP000053097 UP000279307 UP000092461 UP000002282 UP000008744 UP000007798 UP000001819 UP000075809 UP000001070 UP000268350 UP000008237 UP000007819 UP000092553 UP000009192
Interpro
IPR011001
Saposin-like
+ More
IPR041805 ASMase/PPN1_MPP
IPR029052 Metallo-depent_PP-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR008139 SaposinB_dom
IPR011160 Sphingomy_PDE
IPR017853 Glycoside_hydrolase_SF
IPR006047 Glyco_hydro_13_cat_dom
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR007856 SapB_1
IPR041805 ASMase/PPN1_MPP
IPR029052 Metallo-depent_PP-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR008139 SaposinB_dom
IPR011160 Sphingomy_PDE
IPR017853 Glycoside_hydrolase_SF
IPR006047 Glyco_hydro_13_cat_dom
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR007856 SapB_1
Gene 3D
ProteinModelPortal
A0A2H1WW48
A0A194RHF1
A0A194PN53
I4DM60
A0A212FIX9
A0A182U186
+ More
A0A182XJQ6 A0A182PMW3 A0A182VGX6 A0A182L586 A0A182IIK0 A0A182Q6E2 A0A182IIK1 Q7PZJ1 A0A182Y9I9 A0A182KEX5 A0A182M166 A0A084WIG8 A0A182RXV4 A0A182JHX6 A0A182N2R8 A0A182WQP9 A0A182SBY3 A0A182RXV5 A0A182WQP8 A0A1L8DPQ0 A0A1L8DQ04 A0A1L8DPS5 A0A1S4F4J5 Q17FG9 W5JWZ2 A0A182FZW9 Q17FG8 A0A182FZW8 Q17FH0 W8BSV3 W8ATX5 A0A2M4ALK1 A0A2M4ANM1 A0A2M4BFI8 W8BG77 A0A2M4ALB3 A0A2M4ALQ8 A0A2M4BGF7 A0A2K6VB97 A0A034V391 A0A0K8UKX6 D6WVW3 A0A139WDF2 A0A0A1XFR8 A0A0A1XCZ7 A0A0K8VNE0 A0A034V153 A0A0K8U320 B0WS01 A0A0T6B856 A0A1A9ZW88 A0A1B0FF57 U5TZ69 A0A1Q3FC40 A0A1Q3FEJ9 A0A1Q3FEE2 A0A1Q3FD90 A0A1Q3FBV2 A0A1I8P9V5 A0A1I8P9N2 A0A1I8P9M7 D3TNH1 A0A1A9UKZ8 A0A1A9X4A2 A0A1B0BYE8 A0A1W4UZV4 A0A1A9YGB8 A0A1W4V1F0 A0A0J9RJW2 A0A0J9UA65 A0A0J9RKB0 A0A0J9RKG8 B4QBX0 B3NQH4 A0A336MAU4 A0A026WT08 A0A336MIZ9 A0A1B0GGU0 B4PAW9 B4H694 A0A0R1DTP2 B4MYD9 Q28WU3 N6VFZ6 A0A151WVL7 B4J4T2 A0A3B0IYW2 A0A3B0J2R2 E2C0J8 A0A3B0IZ62 J9JZU0 A0A0M4EDA7 A0A0Q9XK69 B4KMI1
A0A182XJQ6 A0A182PMW3 A0A182VGX6 A0A182L586 A0A182IIK0 A0A182Q6E2 A0A182IIK1 Q7PZJ1 A0A182Y9I9 A0A182KEX5 A0A182M166 A0A084WIG8 A0A182RXV4 A0A182JHX6 A0A182N2R8 A0A182WQP9 A0A182SBY3 A0A182RXV5 A0A182WQP8 A0A1L8DPQ0 A0A1L8DQ04 A0A1L8DPS5 A0A1S4F4J5 Q17FG9 W5JWZ2 A0A182FZW9 Q17FG8 A0A182FZW8 Q17FH0 W8BSV3 W8ATX5 A0A2M4ALK1 A0A2M4ANM1 A0A2M4BFI8 W8BG77 A0A2M4ALB3 A0A2M4ALQ8 A0A2M4BGF7 A0A2K6VB97 A0A034V391 A0A0K8UKX6 D6WVW3 A0A139WDF2 A0A0A1XFR8 A0A0A1XCZ7 A0A0K8VNE0 A0A034V153 A0A0K8U320 B0WS01 A0A0T6B856 A0A1A9ZW88 A0A1B0FF57 U5TZ69 A0A1Q3FC40 A0A1Q3FEJ9 A0A1Q3FEE2 A0A1Q3FD90 A0A1Q3FBV2 A0A1I8P9V5 A0A1I8P9N2 A0A1I8P9M7 D3TNH1 A0A1A9UKZ8 A0A1A9X4A2 A0A1B0BYE8 A0A1W4UZV4 A0A1A9YGB8 A0A1W4V1F0 A0A0J9RJW2 A0A0J9UA65 A0A0J9RKB0 A0A0J9RKG8 B4QBX0 B3NQH4 A0A336MAU4 A0A026WT08 A0A336MIZ9 A0A1B0GGU0 B4PAW9 B4H694 A0A0R1DTP2 B4MYD9 Q28WU3 N6VFZ6 A0A151WVL7 B4J4T2 A0A3B0IYW2 A0A3B0J2R2 E2C0J8 A0A3B0IZ62 J9JZU0 A0A0M4EDA7 A0A0Q9XK69 B4KMI1
PDB
5FIC
E-value=1.02475e-58,
Score=575
Ontologies
PATHWAY
GO
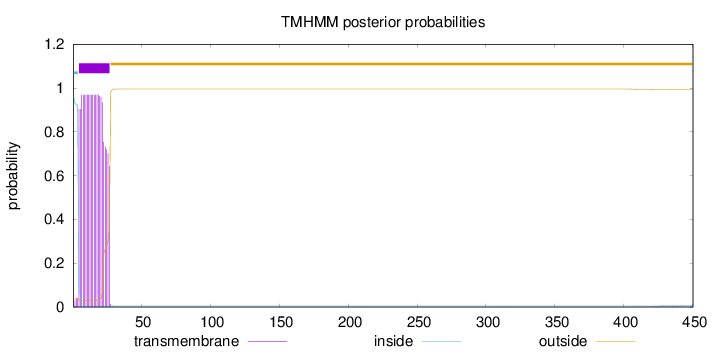
Topology
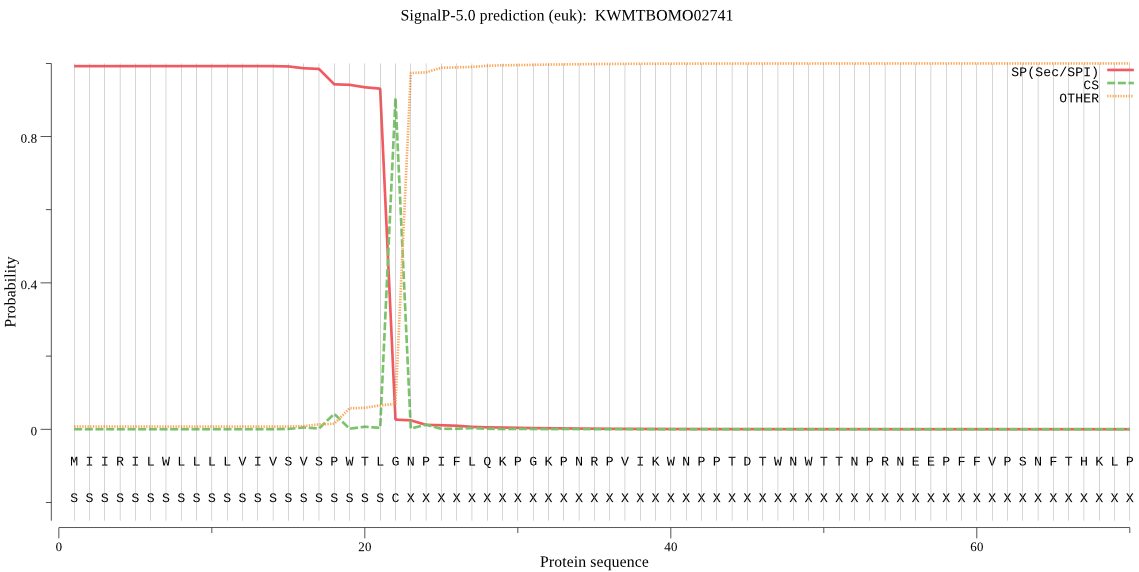
SignalP
Position: 1 - 22,
Likelihood: 0.992230
Length:
450
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.98943
Exp number, first 60 AAs:
20.93717
Total prob of N-in:
0.96377
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 450
Population Genetic Test Statistics
Pi
203.396761
Theta
171.515688
Tajima's D
1.131154
CLR
0.136266
CSRT
0.695065246737663
Interpretation
Uncertain
