Gene
KWMTBOMO02740
Pre Gene Modal
BGIBMGA003720
Annotation
PREDICTED:_sphingomyelin_phosphodiesterase_isoform_X1_[Bombyx_mori]
Full name
Sphingomyelin phosphodiesterase
Location in the cell
Mitochondrial Reliability : 2.252
Sequence
CDS
ATGAATTTGAAAGAAGCAAATCTGTTTGGATATCCGATATGGTATAAGTTGTACTCGGCCAGGTCTGCGTACCTGATGCCGGCGTTGCGACCGCAAGACTGGGACAAGTTTATTGACGACATGACCGCGAAAGAAGAAGTTTTCAATTTGTATTACAAACACTATTGGAAGAACAGTCCCCGCAGAGGTATGTGCGACAGTGAATGTCGTAAGCGATTGATTTGCGACGCGCGGTCGGGACGATCACACGACCGAAGGGCCCTCTGCGTGCACATCGAAGCACGCATCGACGGCGCCGCACCCACTCCACAGACTTGGAGAGCTTGGCTATATAATGGCTTGTCTGTTTCAGATGAACCTAGCGTACAACAAGTATAA
Protein
MNLKEANLFGYPIWYKLYSARSAYLMPALRPQDWDKFIDDMTAKEEVFNLYYKHYWKNSPRRGMCDSECRKRLICDARSGRSHDRRALCVHIEARIDGAAPTPQTWRAWLYNGLSVSDEPSVQQV
Summary
Description
Converts sphingomyelin to ceramide.
Catalytic Activity
H2O + sphingomyelin = an N-acylsphing-4-enine + H(+) + phosphocholine
Cofactor
Zn(2+)
Similarity
Belongs to the acid sphingomyelinase family.
Feature
chain Sphingomyelin phosphodiesterase
Uniprot
H9J2I3
I4DNR9
A0A2H1WW48
A0A212FIX9
I4DM60
A0A194RHF1
+ More
A0A0L7KM87 A0A1E1WJC8 A0A194PN53 A0A2P8Y985 A0A151JNZ1 A0A2H8TJK1 A0A1B6CHV1 A0A1B6BX76 A0A0C9R1P7 A0A2S2NPD7 A0A0M8ZR76 A0A0A1XCZ7 A0A2J7Q8U9 A0A2J7Q8T3 A0A2J7Q8T9 A0A154PI80 A0A2S2QDZ0 A0A067RKN2 J9JZU0 A0A0Q9WFK0 A0A1I8P9M7 A0A0A1XFR8 B4J4T2 A0A151JWZ9 B4LK49 A0A151WVL7 A0A195D6Z5 A0A158NFM2 A0A1A9X4A2 F4X0C3 A0A195AXW9 A0A0T6B7T0 A0A1B6F6D2 B4MYD9 A0A1A9UKZ8 A0A0Q9XK69 A0A1I8MSL3 A0A0M4EDA7 B3MDM0 T1P878 T1PA28 B4KMI1 A0A0J7L1C1 A0A1I8MSL0 A0A1B6L7T1 A0A1I8MSI9 A0A1I8P9V5 A0A3B0IYW2 A0A1I8P9N2 A0A1I8MSJ2 A0A1A9YGB8 A0A1I8MSL2 A0A0K8VNE0 A0A026WT08 A0A1B6MBG3 A0A3B0IZ62 A0A3B0J2R2 A0A336MIZ9 A0A0P8XQL7 A0A1B6FP94 A0A0P9AG89 E9IEU7 A0A1B0BYE8 W8BSV3 A0A1A9ZW88 A0A1B0FF57 B4H693 D3TNH1 E2C0J8 A0A1B6MBI5 A0A088A9Y5 A0A034V391 A0A0K8UKX6 A0A2A3E0P7 A0A0K8U320 A0A034V153 E0VT73 W8BG77 W8ATX5 A0A182KEX5 A0A182JHX6 A0A224XJK0 A0A182Q6E2 A0A182N2R8 A0A023F8R8 A0A1B0GGU0 R4G4X4 A0A1L8DPQ0 A0A1L8DPS5 B4IHE2 A0A1L8DQ04 A0A1Y1LHP2 A0A2M4BGF7
A0A0L7KM87 A0A1E1WJC8 A0A194PN53 A0A2P8Y985 A0A151JNZ1 A0A2H8TJK1 A0A1B6CHV1 A0A1B6BX76 A0A0C9R1P7 A0A2S2NPD7 A0A0M8ZR76 A0A0A1XCZ7 A0A2J7Q8U9 A0A2J7Q8T3 A0A2J7Q8T9 A0A154PI80 A0A2S2QDZ0 A0A067RKN2 J9JZU0 A0A0Q9WFK0 A0A1I8P9M7 A0A0A1XFR8 B4J4T2 A0A151JWZ9 B4LK49 A0A151WVL7 A0A195D6Z5 A0A158NFM2 A0A1A9X4A2 F4X0C3 A0A195AXW9 A0A0T6B7T0 A0A1B6F6D2 B4MYD9 A0A1A9UKZ8 A0A0Q9XK69 A0A1I8MSL3 A0A0M4EDA7 B3MDM0 T1P878 T1PA28 B4KMI1 A0A0J7L1C1 A0A1I8MSL0 A0A1B6L7T1 A0A1I8MSI9 A0A1I8P9V5 A0A3B0IYW2 A0A1I8P9N2 A0A1I8MSJ2 A0A1A9YGB8 A0A1I8MSL2 A0A0K8VNE0 A0A026WT08 A0A1B6MBG3 A0A3B0IZ62 A0A3B0J2R2 A0A336MIZ9 A0A0P8XQL7 A0A1B6FP94 A0A0P9AG89 E9IEU7 A0A1B0BYE8 W8BSV3 A0A1A9ZW88 A0A1B0FF57 B4H693 D3TNH1 E2C0J8 A0A1B6MBI5 A0A088A9Y5 A0A034V391 A0A0K8UKX6 A0A2A3E0P7 A0A0K8U320 A0A034V153 E0VT73 W8BG77 W8ATX5 A0A182KEX5 A0A182JHX6 A0A224XJK0 A0A182Q6E2 A0A182N2R8 A0A023F8R8 A0A1B0GGU0 R4G4X4 A0A1L8DPQ0 A0A1L8DPS5 B4IHE2 A0A1L8DQ04 A0A1Y1LHP2 A0A2M4BGF7
EC Number
3.1.4.12
Pubmed
EMBL
BABH01007983
AK403101
BAM19559.1
ODYU01011505
SOQ57278.1
AGBW02008321
+ More
OWR53682.1 AK402378 BAM19000.1 KQ460207 KPJ16750.1 JTDY01008749 KOB64418.1 GDQN01003935 JAT87119.1 KQ459597 KPI94861.1 PYGN01000786 PSN40825.1 KQ978791 KYN28340.1 GFXV01001653 MBW13458.1 GEDC01024328 JAS12970.1 GEDC01031406 JAS05892.1 GBYB01010059 JAG79826.1 GGMR01006395 MBY19014.1 KQ435936 KOX68326.1 GBXI01005491 JAD08801.1 NEVH01016946 PNF24993.1 PNF24992.1 PNF24994.1 KQ434924 KZC11549.1 GGMS01006698 MBY75901.1 KK852458 KDR23573.1 ABLF02039753 CH940648 KRF79525.1 GBXI01004959 JAD09333.1 CH916367 EDW00628.1 KQ981620 KYN39223.1 EDW60638.2 KQ982699 KYQ51950.1 KQ976750 KYN08647.1 ADTU01014665 ADTU01014666 ADTU01014667 GL888497 EGI60081.1 KQ976716 KYM76887.1 LJIG01009347 KRT83262.1 GECZ01024024 JAS45745.1 CH963894 EDW77128.2 CH933808 KRG05506.1 CP012524 ALC41877.1 CH902619 EDV37483.2 KA644797 AFP59426.1 KA644798 AFP59427.1 EDW10828.1 LBMM01001397 KMQ96388.1 GEBQ01020215 JAT19762.1 OUUW01000001 SPP73394.1 GDHF01011917 JAI40397.1 KK107119 QOIP01000009 EZA58816.1 RLU18737.1 GEBQ01006712 JAT33265.1 SPP73395.1 SPP73393.1 UFQT01000641 SSX26028.1 KPU76858.1 GECZ01017734 JAS52035.1 KPU76857.1 GL762722 EFZ20884.1 JXJN01022601 GAMC01014261 JAB92294.1 CCAG010006382 CCAG010006383 CH479213 EDW33317.1 EZ422973 JQ308535 ADD19249.1 AFJ68090.1 GL451800 EFN78515.1 GEBQ01006685 JAT33292.1 GAKP01021948 JAC37004.1 GDHF01024977 JAI27337.1 KZ288472 PBC25325.1 GDHF01031381 JAI20933.1 GAKP01021946 JAC37006.1 DS235760 EEB16579.1 GAMC01014259 JAB92296.1 GAMC01014260 JAB92295.1 GFTR01007786 JAW08640.1 AXCN02001087 GBBI01000925 JAC17787.1 AJWK01000745 AJWK01000746 AJWK01000747 ACPB03007771 GAHY01000995 JAA76515.1 GFDF01005626 JAV08458.1 GFDF01005628 JAV08456.1 CH480838 EDW49166.1 GFDF01005627 JAV08457.1 GEZM01055093 GEZM01055092 JAV73154.1 GGFJ01002747 MBW51888.1
OWR53682.1 AK402378 BAM19000.1 KQ460207 KPJ16750.1 JTDY01008749 KOB64418.1 GDQN01003935 JAT87119.1 KQ459597 KPI94861.1 PYGN01000786 PSN40825.1 KQ978791 KYN28340.1 GFXV01001653 MBW13458.1 GEDC01024328 JAS12970.1 GEDC01031406 JAS05892.1 GBYB01010059 JAG79826.1 GGMR01006395 MBY19014.1 KQ435936 KOX68326.1 GBXI01005491 JAD08801.1 NEVH01016946 PNF24993.1 PNF24992.1 PNF24994.1 KQ434924 KZC11549.1 GGMS01006698 MBY75901.1 KK852458 KDR23573.1 ABLF02039753 CH940648 KRF79525.1 GBXI01004959 JAD09333.1 CH916367 EDW00628.1 KQ981620 KYN39223.1 EDW60638.2 KQ982699 KYQ51950.1 KQ976750 KYN08647.1 ADTU01014665 ADTU01014666 ADTU01014667 GL888497 EGI60081.1 KQ976716 KYM76887.1 LJIG01009347 KRT83262.1 GECZ01024024 JAS45745.1 CH963894 EDW77128.2 CH933808 KRG05506.1 CP012524 ALC41877.1 CH902619 EDV37483.2 KA644797 AFP59426.1 KA644798 AFP59427.1 EDW10828.1 LBMM01001397 KMQ96388.1 GEBQ01020215 JAT19762.1 OUUW01000001 SPP73394.1 GDHF01011917 JAI40397.1 KK107119 QOIP01000009 EZA58816.1 RLU18737.1 GEBQ01006712 JAT33265.1 SPP73395.1 SPP73393.1 UFQT01000641 SSX26028.1 KPU76858.1 GECZ01017734 JAS52035.1 KPU76857.1 GL762722 EFZ20884.1 JXJN01022601 GAMC01014261 JAB92294.1 CCAG010006382 CCAG010006383 CH479213 EDW33317.1 EZ422973 JQ308535 ADD19249.1 AFJ68090.1 GL451800 EFN78515.1 GEBQ01006685 JAT33292.1 GAKP01021948 JAC37004.1 GDHF01024977 JAI27337.1 KZ288472 PBC25325.1 GDHF01031381 JAI20933.1 GAKP01021946 JAC37006.1 DS235760 EEB16579.1 GAMC01014259 JAB92296.1 GAMC01014260 JAB92295.1 GFTR01007786 JAW08640.1 AXCN02001087 GBBI01000925 JAC17787.1 AJWK01000745 AJWK01000746 AJWK01000747 ACPB03007771 GAHY01000995 JAA76515.1 GFDF01005626 JAV08458.1 GFDF01005628 JAV08456.1 CH480838 EDW49166.1 GFDF01005627 JAV08457.1 GEZM01055093 GEZM01055092 JAV73154.1 GGFJ01002747 MBW51888.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000053268
UP000245037
+ More
UP000078492 UP000053105 UP000235965 UP000076502 UP000027135 UP000007819 UP000008792 UP000095300 UP000001070 UP000078541 UP000075809 UP000078542 UP000005205 UP000091820 UP000007755 UP000078540 UP000007798 UP000078200 UP000009192 UP000095301 UP000092553 UP000007801 UP000036403 UP000268350 UP000092443 UP000053097 UP000279307 UP000092460 UP000092445 UP000092444 UP000008744 UP000008237 UP000005203 UP000242457 UP000009046 UP000075881 UP000075880 UP000075886 UP000075884 UP000092461 UP000015103 UP000001292
UP000078492 UP000053105 UP000235965 UP000076502 UP000027135 UP000007819 UP000008792 UP000095300 UP000001070 UP000078541 UP000075809 UP000078542 UP000005205 UP000091820 UP000007755 UP000078540 UP000007798 UP000078200 UP000009192 UP000095301 UP000092553 UP000007801 UP000036403 UP000268350 UP000092443 UP000053097 UP000279307 UP000092460 UP000092445 UP000092444 UP000008744 UP000008237 UP000005203 UP000242457 UP000009046 UP000075881 UP000075880 UP000075886 UP000075884 UP000092461 UP000015103 UP000001292
PRIDE
Interpro
SUPFAM
SSF47862
SSF47862
Gene 3D
ProteinModelPortal
H9J2I3
I4DNR9
A0A2H1WW48
A0A212FIX9
I4DM60
A0A194RHF1
+ More
A0A0L7KM87 A0A1E1WJC8 A0A194PN53 A0A2P8Y985 A0A151JNZ1 A0A2H8TJK1 A0A1B6CHV1 A0A1B6BX76 A0A0C9R1P7 A0A2S2NPD7 A0A0M8ZR76 A0A0A1XCZ7 A0A2J7Q8U9 A0A2J7Q8T3 A0A2J7Q8T9 A0A154PI80 A0A2S2QDZ0 A0A067RKN2 J9JZU0 A0A0Q9WFK0 A0A1I8P9M7 A0A0A1XFR8 B4J4T2 A0A151JWZ9 B4LK49 A0A151WVL7 A0A195D6Z5 A0A158NFM2 A0A1A9X4A2 F4X0C3 A0A195AXW9 A0A0T6B7T0 A0A1B6F6D2 B4MYD9 A0A1A9UKZ8 A0A0Q9XK69 A0A1I8MSL3 A0A0M4EDA7 B3MDM0 T1P878 T1PA28 B4KMI1 A0A0J7L1C1 A0A1I8MSL0 A0A1B6L7T1 A0A1I8MSI9 A0A1I8P9V5 A0A3B0IYW2 A0A1I8P9N2 A0A1I8MSJ2 A0A1A9YGB8 A0A1I8MSL2 A0A0K8VNE0 A0A026WT08 A0A1B6MBG3 A0A3B0IZ62 A0A3B0J2R2 A0A336MIZ9 A0A0P8XQL7 A0A1B6FP94 A0A0P9AG89 E9IEU7 A0A1B0BYE8 W8BSV3 A0A1A9ZW88 A0A1B0FF57 B4H693 D3TNH1 E2C0J8 A0A1B6MBI5 A0A088A9Y5 A0A034V391 A0A0K8UKX6 A0A2A3E0P7 A0A0K8U320 A0A034V153 E0VT73 W8BG77 W8ATX5 A0A182KEX5 A0A182JHX6 A0A224XJK0 A0A182Q6E2 A0A182N2R8 A0A023F8R8 A0A1B0GGU0 R4G4X4 A0A1L8DPQ0 A0A1L8DPS5 B4IHE2 A0A1L8DQ04 A0A1Y1LHP2 A0A2M4BGF7
A0A0L7KM87 A0A1E1WJC8 A0A194PN53 A0A2P8Y985 A0A151JNZ1 A0A2H8TJK1 A0A1B6CHV1 A0A1B6BX76 A0A0C9R1P7 A0A2S2NPD7 A0A0M8ZR76 A0A0A1XCZ7 A0A2J7Q8U9 A0A2J7Q8T3 A0A2J7Q8T9 A0A154PI80 A0A2S2QDZ0 A0A067RKN2 J9JZU0 A0A0Q9WFK0 A0A1I8P9M7 A0A0A1XFR8 B4J4T2 A0A151JWZ9 B4LK49 A0A151WVL7 A0A195D6Z5 A0A158NFM2 A0A1A9X4A2 F4X0C3 A0A195AXW9 A0A0T6B7T0 A0A1B6F6D2 B4MYD9 A0A1A9UKZ8 A0A0Q9XK69 A0A1I8MSL3 A0A0M4EDA7 B3MDM0 T1P878 T1PA28 B4KMI1 A0A0J7L1C1 A0A1I8MSL0 A0A1B6L7T1 A0A1I8MSI9 A0A1I8P9V5 A0A3B0IYW2 A0A1I8P9N2 A0A1I8MSJ2 A0A1A9YGB8 A0A1I8MSL2 A0A0K8VNE0 A0A026WT08 A0A1B6MBG3 A0A3B0IZ62 A0A3B0J2R2 A0A336MIZ9 A0A0P8XQL7 A0A1B6FP94 A0A0P9AG89 E9IEU7 A0A1B0BYE8 W8BSV3 A0A1A9ZW88 A0A1B0FF57 B4H693 D3TNH1 E2C0J8 A0A1B6MBI5 A0A088A9Y5 A0A034V391 A0A0K8UKX6 A0A2A3E0P7 A0A0K8U320 A0A034V153 E0VT73 W8BG77 W8ATX5 A0A182KEX5 A0A182JHX6 A0A224XJK0 A0A182Q6E2 A0A182N2R8 A0A023F8R8 A0A1B0GGU0 R4G4X4 A0A1L8DPQ0 A0A1L8DPS5 B4IHE2 A0A1L8DQ04 A0A1Y1LHP2 A0A2M4BGF7
PDB
5FIC
E-value=7.19436e-09,
Score=137
Ontologies
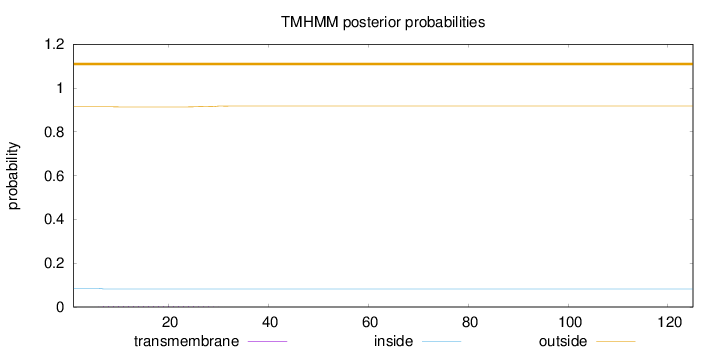
Topology
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08899
Exp number, first 60 AAs:
0.08899
Total prob of N-in:
0.08587
outside
1 - 125
Population Genetic Test Statistics
Pi
243.560594
Theta
177.252798
Tajima's D
1.202218
CLR
0.041257
CSRT
0.711614419279036
Interpretation
Uncertain
