Pre Gene Modal
BGIBMGA003475
Annotation
PREDICTED:_elongation_factor_1_delta_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.485 Nuclear Reliability : 1.278
Sequence
CDS
ATGGATAGTGTAGCAACTTTGGCGGGTGTTCCTAATACAGAGCTCAGTACTAAAGTCATCAGCTTAGAAAACGAAAACAAGGATTTGAAAAAGGCTATAGATGATTTACGTAATTTAGTCATCAGCCTGCAAGTCCGTGTTGAAACCTTGGAATCATCCAGCAGTATCACCTCTGATTGTATTCAATCAGAAAAACCTGCAGCAGCTTCTACTGATAAGGATGACGAAGATGATGATGTTGATTTGTTTGGTTCAGATGATGAAGAGGAGAGTGCTGAAGCAGCTAAGATAAAGGAAGAGCGTCTTGCTGCATATAATGCAAAGAAAGCTAAAAAGCCAGTGCTAATTGCAAAGTCGAATATTATTCTGGATGTAAAGCCATGGGATGATGAGACTGATATGGCAGCCTTGGAACAAGCTGTAAGGTCAATCAGCACTGATGGCCTCTTGTGGGGTGCCGCTAAACTAGTGCCTTTGGCTTATGGTATCCATAAGCTGCAAATTTCCTGTGTTGTTGAAGATGATAAGGTTTCTATTGATTGGCTTACTGAGGAAATCGAGAAAAATGAAGACTATGTACAAAGTGTCGATATTGCTGCGTTCAACAAAGTCTAG
Protein
MDSVATLAGVPNTELSTKVISLENENKDLKKAIDDLRNLVISLQVRVETLESSSSITSDCIQSEKPAAASTDKDDEDDDVDLFGSDDEEESAEAAKIKEERLAAYNAKKAKKPVLIAKSNIILDVKPWDDETDMAALEQAVRSISTDGLLWGAAKLVPLAYGIHKLQISCVVEDDKVSIDWLTEEIEKNEDYVQSVDIAAFNKV
Summary
Similarity
Belongs to the EF-1-beta/EF-1-delta family.
Uniprot
Q9BPS1
A0A212FIY9
A0A0L7LV85
I4DIY3
I4DMJ6
A0A2H1WYK3
+ More
A0A2A4JYE9 A0A2A4JZU8 A0A1E1WTF4 A0A1E1W7W5 A0A1E1VZX5 S4NT30 A0A194RH23 A0A1B0CGT9 A0A1L8DH59 A0A1L8DGI4 A0A182Q2L1 A0A182PD75 E1ZUU8 A0A182Y5E1 A0A182JD47 A0A182MJK1 A0A182RV62 A0A348G620 A0A0K8TQ13 A0A1B6L2A8 Q1W2A4 A0A1B6MBD5 A0A1B6KQ50 A0A1B6MTR1 A0A151JY62 A0A158NB19 A0A182ND03 A0A151I7W1 A0A195DYN9 A0A0P8XGN7 B3MPN3 A0A2S2QDE6 U5EXR8 T1JB50 A0A1B6CX42 A0A0P6ES73 A0A0P5UXS2 A0A0P5A6D7 A0A0N8E5Y1 A0A0P5G0H4 A0A0P5L1D4 A0A0N8D0Z3 A0A0P5YHD9 A0A0P5I1K7 A0A0P5ERM8 A0A182SIU4
A0A2A4JYE9 A0A2A4JZU8 A0A1E1WTF4 A0A1E1W7W5 A0A1E1VZX5 S4NT30 A0A194RH23 A0A1B0CGT9 A0A1L8DH59 A0A1L8DGI4 A0A182Q2L1 A0A182PD75 E1ZUU8 A0A182Y5E1 A0A182JD47 A0A182MJK1 A0A182RV62 A0A348G620 A0A0K8TQ13 A0A1B6L2A8 Q1W2A4 A0A1B6MBD5 A0A1B6KQ50 A0A1B6MTR1 A0A151JY62 A0A158NB19 A0A182ND03 A0A151I7W1 A0A195DYN9 A0A0P8XGN7 B3MPN3 A0A2S2QDE6 U5EXR8 T1JB50 A0A1B6CX42 A0A0P6ES73 A0A0P5UXS2 A0A0P5A6D7 A0A0N8E5Y1 A0A0P5G0H4 A0A0P5L1D4 A0A0N8D0Z3 A0A0P5YHD9 A0A0P5I1K7 A0A0P5ERM8 A0A182SIU4
Pubmed
EMBL
BABH01007991
AB046366
BAB21109.1
AGBW02008321
OWR53691.1
JTDY01000022
+ More
KOB79372.1 AK401251 KQ459597 BAM17873.1 KPI94852.1 AK402514 BAM19136.1 ODYU01011613 SOQ57464.1 NWSH01000357 PCG77045.1 PCG77043.1 GDQN01000800 JAT90254.1 GDQN01008017 JAT83037.1 GDQN01010843 JAT80211.1 GAIX01012336 JAA80224.1 KQ460207 KPJ16739.1 AJWK01011585 AJWK01011586 AJWK01011587 GFDF01008359 JAV05725.1 GFDF01008640 JAV05444.1 AXCN02001566 GL434297 EFN75040.1 AXCM01001325 FX985559 BBF97893.1 GDAI01001372 JAI16231.1 GEBQ01022115 JAT17862.1 DQ445523 GEBQ01032049 ABD98761.1 JAT07928.1 GEBQ01006749 JAT33228.1 GEBQ01026633 JAT13344.1 GEBQ01000708 JAT39269.1 KQ981533 KYN40114.1 ADTU01010746 KQ978396 KYM94224.1 KQ980050 KYN18015.1 CH902620 KPU73879.1 EDV32281.1 GGMS01006546 MBY75749.1 GANO01000805 JAB59066.1 JH432008 GEDC01019307 JAS17991.1 GDIQ01058621 JAN36116.1 GDIP01110053 GDIP01110052 JAL93661.1 GDIP01207848 JAJ15554.1 GDIQ01058620 JAN36117.1 GDIQ01249261 JAK02464.1 GDIQ01177632 GDIQ01177574 JAK74151.1 GDIP01079503 JAM24212.1 GDIP01071570 JAM32145.1 GDIQ01221920 JAK29805.1 GDIQ01265994 JAJ85730.1
KOB79372.1 AK401251 KQ459597 BAM17873.1 KPI94852.1 AK402514 BAM19136.1 ODYU01011613 SOQ57464.1 NWSH01000357 PCG77045.1 PCG77043.1 GDQN01000800 JAT90254.1 GDQN01008017 JAT83037.1 GDQN01010843 JAT80211.1 GAIX01012336 JAA80224.1 KQ460207 KPJ16739.1 AJWK01011585 AJWK01011586 AJWK01011587 GFDF01008359 JAV05725.1 GFDF01008640 JAV05444.1 AXCN02001566 GL434297 EFN75040.1 AXCM01001325 FX985559 BBF97893.1 GDAI01001372 JAI16231.1 GEBQ01022115 JAT17862.1 DQ445523 GEBQ01032049 ABD98761.1 JAT07928.1 GEBQ01006749 JAT33228.1 GEBQ01026633 JAT13344.1 GEBQ01000708 JAT39269.1 KQ981533 KYN40114.1 ADTU01010746 KQ978396 KYM94224.1 KQ980050 KYN18015.1 CH902620 KPU73879.1 EDV32281.1 GGMS01006546 MBY75749.1 GANO01000805 JAB59066.1 JH432008 GEDC01019307 JAS17991.1 GDIQ01058621 JAN36116.1 GDIP01110053 GDIP01110052 JAL93661.1 GDIP01207848 JAJ15554.1 GDIQ01058620 JAN36117.1 GDIQ01249261 JAK02464.1 GDIQ01177632 GDIQ01177574 JAK74151.1 GDIP01079503 JAM24212.1 GDIP01071570 JAM32145.1 GDIQ01221920 JAK29805.1 GDIQ01265994 JAJ85730.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF54984
SSF54984
Gene 3D
CDD
ProteinModelPortal
Q9BPS1
A0A212FIY9
A0A0L7LV85
I4DIY3
I4DMJ6
A0A2H1WYK3
+ More
A0A2A4JYE9 A0A2A4JZU8 A0A1E1WTF4 A0A1E1W7W5 A0A1E1VZX5 S4NT30 A0A194RH23 A0A1B0CGT9 A0A1L8DH59 A0A1L8DGI4 A0A182Q2L1 A0A182PD75 E1ZUU8 A0A182Y5E1 A0A182JD47 A0A182MJK1 A0A182RV62 A0A348G620 A0A0K8TQ13 A0A1B6L2A8 Q1W2A4 A0A1B6MBD5 A0A1B6KQ50 A0A1B6MTR1 A0A151JY62 A0A158NB19 A0A182ND03 A0A151I7W1 A0A195DYN9 A0A0P8XGN7 B3MPN3 A0A2S2QDE6 U5EXR8 T1JB50 A0A1B6CX42 A0A0P6ES73 A0A0P5UXS2 A0A0P5A6D7 A0A0N8E5Y1 A0A0P5G0H4 A0A0P5L1D4 A0A0N8D0Z3 A0A0P5YHD9 A0A0P5I1K7 A0A0P5ERM8 A0A182SIU4
A0A2A4JYE9 A0A2A4JZU8 A0A1E1WTF4 A0A1E1W7W5 A0A1E1VZX5 S4NT30 A0A194RH23 A0A1B0CGT9 A0A1L8DH59 A0A1L8DGI4 A0A182Q2L1 A0A182PD75 E1ZUU8 A0A182Y5E1 A0A182JD47 A0A182MJK1 A0A182RV62 A0A348G620 A0A0K8TQ13 A0A1B6L2A8 Q1W2A4 A0A1B6MBD5 A0A1B6KQ50 A0A1B6MTR1 A0A151JY62 A0A158NB19 A0A182ND03 A0A151I7W1 A0A195DYN9 A0A0P8XGN7 B3MPN3 A0A2S2QDE6 U5EXR8 T1JB50 A0A1B6CX42 A0A0P6ES73 A0A0P5UXS2 A0A0P5A6D7 A0A0N8E5Y1 A0A0P5G0H4 A0A0P5L1D4 A0A0N8D0Z3 A0A0P5YHD9 A0A0P5I1K7 A0A0P5ERM8 A0A182SIU4
PDB
2N51
E-value=1.36779e-33,
Score=354
Ontologies
GO
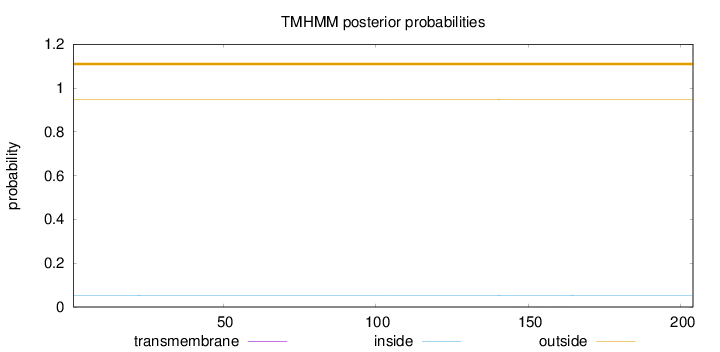
Topology
Length:
204
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00808
Exp number, first 60 AAs:
0.00073
Total prob of N-in:
0.05285
outside
1 - 204
Population Genetic Test Statistics
Pi
198.018744
Theta
154.95465
Tajima's D
0.302511
CLR
0.227714
CSRT
0.460076996150192
Interpretation
Uncertain
