Gene
KWMTBOMO02733
Annotation
PREDICTED:_cytoplasmic_tRNA_2-thiolation_protein_2_[Amyelois_transitella]
Full name
Cytoplasmic tRNA 2-thiolation protein 2
Location in the cell
Nuclear Reliability : 3.18
Sequence
CDS
ATGAAATGTAAAAAATGCAATTCATCTTCTGCATTAATTATCTTAAGAAAAAAAGATTTGTATTGCAGAGAGTGCTTCCTAGCAAATGTAACTCATAAATTCCGTTCTAGCATCGGAAAAAATAAAATATTAGGAAAGGCAGATAATGTTTTGATCTGTTTGTCGGGGAAGGAAGGCTCAACGGTTTTATTAGATTTGATTAATAATGCGTTATCCCTTGATAACCACAAGAAACTACGAATAACTCCAATTTTTCTACACTTGTGTGAAAACTACGAAACAATAAAGAATGTAATAGAACAATGTCAAAAGTACAATTATGACCTATATTCACCACATATATCAGAATATGTTCAATCCAAATCTCATATACCAATTGTGAATTCTTTACCTATTGTAAACAATGAACTTATAGAAGAGTACAATAACATGCAAAATAGCATGTTACCAACAGTTAAAGATGATTTCATAACTAATATTAAGAGAACTTTATATATTAGATATGCAAAGCTTTTGAATTGCAAATATATTTTCACTGGTGAAACAACAACCACTTTGGCCACAAAATTACTTGTAAATTTAGCTATTGGACGTGGCTCTCAAGTACAAAGTGATGTGGGATTTTTAGACAGTAGAGATATGGAAATCAAAATTATTAGACCTATGAAAGACATAAGTGAAGAGGAATTAAAACTTTATATCAACTCTAAGAATCTTAAAGTAAATGATAAACAAACATCCAAAGAGAATGAGCACACTAGCTTGCAAACAGTAATTAGTACTTTTGTAAGCAGTTTGCAAGAAAATTTTCCTTCCACAATTTCAACAATATGTAAAACTGCTGATAAAATTGGATTACAAGATGAATGTGACAATGAAAGGCATAATATGTGTGTTATTTGTCAAAGCATCTTGAAACCTGATGAGAAAGCAATCTCAGCTTTAGATGCAACAAGTATTTCCAAAATAGTTTCATTGAGAAAACATACAGACTCTGCTATAGATGTGGACATGGAGAAAAATGTTATTAACTCAACAATATTTCCTTATATTCATAATCGTCTATGTTATGCTTGCAGTAGAAATTATGCAGAAATTAAACAAAGCAAAATACCAAGTTACATTATCGAAACACAAAAATAA
Protein
MKCKKCNSSSALIILRKKDLYCRECFLANVTHKFRSSIGKNKILGKADNVLICLSGKEGSTVLLDLINNALSLDNHKKLRITPIFLHLCENYETIKNVIEQCQKYNYDLYSPHISEYVQSKSHIPIVNSLPIVNNELIEEYNNMQNSMLPTVKDDFITNIKRTLYIRYAKLLNCKYIFTGETTTTLATKLLVNLAIGRGSQVQSDVGFLDSRDMEIKIIRPMKDISEEELKLYINSKNLKVNDKQTSKENEHTSLQTVISTFVSSLQENFPSTISTICKTADKIGLQDECDNERHNMCVICQSILKPDEKAISALDATSISKIVSLRKHTDSAIDVDMEKNVINSTIFPYIHNRLCYACSRNYAEIKQSKIPSYIIETQK
Summary
Description
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of tRNA(Lys), tRNA(Glu) and tRNA(Gln). May act by forming a heterodimer with NCS6/CTU1 that ligates sulfur from thiocarboxylated URM1 onto the uridine of tRNAs at wobble position.
Similarity
Belongs to the CTU2/NCS2 family.
Keywords
Complete proteome
Cytoplasm
Reference proteome
tRNA processing
Feature
chain Cytoplasmic tRNA 2-thiolation protein 2
Uniprot
A0A1E1WG68
A0A2A4JZ41
A0A2H1WWL8
A0A212FIZ1
A0A194RFZ8
A0A0C9RFS3
+ More
U4U4P5 J3JV37 A0A1B6JV90 A0A2P8XGU2 A0A0L7KMA1 D6WYA9 A0A1Y1M3Q4 A0A1B6KH68 A0A067RN66 A0A1B6H2N7 A0A1Y1M405 A0A182H3B6 A0A2K7P687 Q17JB7 A0A293LYM3 A0A158NT32 A0A023GJQ7 V4CK50 T1I792 A0A0P4VUE8 R4FQD1 A0A195BG92 W5JGH8 A0A1S3JE92 A0A0T6AVL9 Q9VIV3 A0A2H8TP24 J9JW98 A0A182RFA1 A0A2R7WS26 A0A1A9V1K9 A0NEF7 A0A182LW56 A0A084W473 A0A1I8MEV7 A0A195C8I0 A0A0V0G776 B4I5W3 A0A0L8HE76 A0A195EFH6 A0A1L8DA21 A0A034VS20 A0A2S2P968 A0A0J9R5L9 B4Q9Z1 A0A182V9Y9 A0A182PHM8 A0A182FHY3 A0A2M4BN88 A0A195EZF8 A0A2M3Z9F5 A0A2R5LK50 A0A182NGW9 A0A182WIH7 A0A023F772 A0A0A1X3V7 A0A182QB04 D3TLU7 B0X911 A0A1W4VER7 A0A182XMW4 A0A3Q0IHQ7 A0A1B0FEC6 A0A1Q3F147 A0A1S3DUI6 A0A182HWS0 A0A1B0A943 A0A1B0C105 B4PAZ6 A0A182Y1A7 A0A2M3Z9D5 A0A151WJD9 B3NM45 A0A210R0K2 A0A026WSU3 T1KDJ5 E9IBE4 F4WG48 B3MN57 A0A2M3ZA98 A0A0K8TW66 B4JCV8 U5EXQ1 A0A0K2T6Y0 A0A1I8NMV7 A0A2G8JTY6 B4MYR2 B4M8J8 E9G223 B4KIW3 R7U248 W8BUL7
U4U4P5 J3JV37 A0A1B6JV90 A0A2P8XGU2 A0A0L7KMA1 D6WYA9 A0A1Y1M3Q4 A0A1B6KH68 A0A067RN66 A0A1B6H2N7 A0A1Y1M405 A0A182H3B6 A0A2K7P687 Q17JB7 A0A293LYM3 A0A158NT32 A0A023GJQ7 V4CK50 T1I792 A0A0P4VUE8 R4FQD1 A0A195BG92 W5JGH8 A0A1S3JE92 A0A0T6AVL9 Q9VIV3 A0A2H8TP24 J9JW98 A0A182RFA1 A0A2R7WS26 A0A1A9V1K9 A0NEF7 A0A182LW56 A0A084W473 A0A1I8MEV7 A0A195C8I0 A0A0V0G776 B4I5W3 A0A0L8HE76 A0A195EFH6 A0A1L8DA21 A0A034VS20 A0A2S2P968 A0A0J9R5L9 B4Q9Z1 A0A182V9Y9 A0A182PHM8 A0A182FHY3 A0A2M4BN88 A0A195EZF8 A0A2M3Z9F5 A0A2R5LK50 A0A182NGW9 A0A182WIH7 A0A023F772 A0A0A1X3V7 A0A182QB04 D3TLU7 B0X911 A0A1W4VER7 A0A182XMW4 A0A3Q0IHQ7 A0A1B0FEC6 A0A1Q3F147 A0A1S3DUI6 A0A182HWS0 A0A1B0A943 A0A1B0C105 B4PAZ6 A0A182Y1A7 A0A2M3Z9D5 A0A151WJD9 B3NM45 A0A210R0K2 A0A026WSU3 T1KDJ5 E9IBE4 F4WG48 B3MN57 A0A2M3ZA98 A0A0K8TW66 B4JCV8 U5EXQ1 A0A0K2T6Y0 A0A1I8NMV7 A0A2G8JTY6 B4MYR2 B4M8J8 E9G223 B4KIW3 R7U248 W8BUL7
Pubmed
22118469
26354079
23537049
22516182
29403074
26227816
+ More
18362917 19820115 28004739 24845553 26483478 17510324 21347285 23254933 27129103 20920257 23761445 10731132 12537572 12537569 12364791 24438588 25315136 17994087 25348373 22936249 25474469 25830018 20353571 25244985 28812685 24508170 30249741 21282665 21719571 29023486 21292972 24495485
18362917 19820115 28004739 24845553 26483478 17510324 21347285 23254933 27129103 20920257 23761445 10731132 12537572 12537569 12364791 24438588 25315136 17994087 25348373 22936249 25474469 25830018 20353571 25244985 28812685 24508170 30249741 21282665 21719571 29023486 21292972 24495485
EMBL
GDQN01005071
JAT85983.1
NWSH01000357
PCG77046.1
ODYU01011613
SOQ57465.1
+ More
AGBW02008321 OWR53692.1 KQ460207 KPJ16738.1 GBYB01007215 JAG76982.1 KB632100 ERL88844.1 APGK01041758 BT127103 KB740997 AEE62065.1 ENN75847.1 GECU01004578 JAT03129.1 PYGN01002190 PSN31223.1 JTDY01009137 KOB64059.1 KQ971356 EFA09124.2 GEZM01041407 JAV80439.1 GEBQ01029212 JAT10765.1 KK852408 KDR24498.1 GECZ01000805 JAS68964.1 GEZM01041403 JAV80443.1 JXUM01024467 KQ560691 KXJ81259.1 CH477233 GFWV01009084 MAA33813.1 ADTU01025481 GBBM01001236 JAC34182.1 KB200170 ESP02605.1 ACPB03016783 GDKW01000287 JAI56308.1 GAHY01000464 JAA77046.1 KQ976488 KYM83612.1 ADMH02001577 ETN62015.1 LJIG01022687 KRT79248.1 AE014134 BT046166 AY118308 GFXV01004149 MBW15954.1 ABLF02039905 KK855397 PTY22368.1 AAAB01008960 AXCM01002623 ATLV01020284 KE525297 KFB45017.1 KQ978205 KYM96481.1 GECL01002270 JAP03854.1 CH480822 KQ418417 KOF87419.1 KQ978957 KYN26998.1 GFDF01010776 JAV03308.1 GAKP01014600 GAKP01014599 JAC44353.1 GGMR01013119 MBY25738.1 CM002910 KMY91024.1 CM000361 GGFJ01005323 MBW54464.1 KQ981897 KYN33685.1 GGFM01004381 MBW25132.1 GGLE01005765 MBY09891.1 GBBI01001622 JAC17090.1 GBXI01008857 JAD05435.1 AXCN02000361 EZ422399 ADD18675.1 DS232513 CCAG010022386 GFDL01013764 JAV21281.1 APCN01001535 JXJN01023850 CM000158 GGFM01004395 MBW25146.1 KQ983039 KYQ47920.1 CH954179 NEDP02000981 OWF54553.1 KK107109 QOIP01000008 EZA59053.1 RLU19939.1 CAEY01002017 GL762111 EFZ22034.1 GL888128 EGI66815.1 CH902620 GGFM01004702 MBW25453.1 GDHF01034014 GDHF01029495 JAI18300.1 JAI22819.1 CH916368 GANO01000837 JAB59034.1 HACA01004402 CDW21763.1 MRZV01001265 PIK39188.1 CH963913 CH940654 GL732530 EFX86173.1 CH933807 AMQN01009843 KB306344 ELT99947.1 GAMC01003658 JAC02898.1
AGBW02008321 OWR53692.1 KQ460207 KPJ16738.1 GBYB01007215 JAG76982.1 KB632100 ERL88844.1 APGK01041758 BT127103 KB740997 AEE62065.1 ENN75847.1 GECU01004578 JAT03129.1 PYGN01002190 PSN31223.1 JTDY01009137 KOB64059.1 KQ971356 EFA09124.2 GEZM01041407 JAV80439.1 GEBQ01029212 JAT10765.1 KK852408 KDR24498.1 GECZ01000805 JAS68964.1 GEZM01041403 JAV80443.1 JXUM01024467 KQ560691 KXJ81259.1 CH477233 GFWV01009084 MAA33813.1 ADTU01025481 GBBM01001236 JAC34182.1 KB200170 ESP02605.1 ACPB03016783 GDKW01000287 JAI56308.1 GAHY01000464 JAA77046.1 KQ976488 KYM83612.1 ADMH02001577 ETN62015.1 LJIG01022687 KRT79248.1 AE014134 BT046166 AY118308 GFXV01004149 MBW15954.1 ABLF02039905 KK855397 PTY22368.1 AAAB01008960 AXCM01002623 ATLV01020284 KE525297 KFB45017.1 KQ978205 KYM96481.1 GECL01002270 JAP03854.1 CH480822 KQ418417 KOF87419.1 KQ978957 KYN26998.1 GFDF01010776 JAV03308.1 GAKP01014600 GAKP01014599 JAC44353.1 GGMR01013119 MBY25738.1 CM002910 KMY91024.1 CM000361 GGFJ01005323 MBW54464.1 KQ981897 KYN33685.1 GGFM01004381 MBW25132.1 GGLE01005765 MBY09891.1 GBBI01001622 JAC17090.1 GBXI01008857 JAD05435.1 AXCN02000361 EZ422399 ADD18675.1 DS232513 CCAG010022386 GFDL01013764 JAV21281.1 APCN01001535 JXJN01023850 CM000158 GGFM01004395 MBW25146.1 KQ983039 KYQ47920.1 CH954179 NEDP02000981 OWF54553.1 KK107109 QOIP01000008 EZA59053.1 RLU19939.1 CAEY01002017 GL762111 EFZ22034.1 GL888128 EGI66815.1 CH902620 GGFM01004702 MBW25453.1 GDHF01034014 GDHF01029495 JAI18300.1 JAI22819.1 CH916368 GANO01000837 JAB59034.1 HACA01004402 CDW21763.1 MRZV01001265 PIK39188.1 CH963913 CH940654 GL732530 EFX86173.1 CH933807 AMQN01009843 KB306344 ELT99947.1 GAMC01003658 JAC02898.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000030742
UP000019118
UP000245037
+ More
UP000037510 UP000007266 UP000027135 UP000069940 UP000249989 UP000008820 UP000005205 UP000030746 UP000015103 UP000078540 UP000000673 UP000085678 UP000000803 UP000007819 UP000075900 UP000078200 UP000007062 UP000075883 UP000030765 UP000095301 UP000078542 UP000001292 UP000053454 UP000078492 UP000000304 UP000075903 UP000075885 UP000069272 UP000078541 UP000075884 UP000075920 UP000075886 UP000002320 UP000192221 UP000076407 UP000079169 UP000092444 UP000075840 UP000092445 UP000092460 UP000002282 UP000076408 UP000075809 UP000008711 UP000242188 UP000053097 UP000279307 UP000015104 UP000007755 UP000007801 UP000001070 UP000095300 UP000230750 UP000007798 UP000008792 UP000000305 UP000009192 UP000014760
UP000037510 UP000007266 UP000027135 UP000069940 UP000249989 UP000008820 UP000005205 UP000030746 UP000015103 UP000078540 UP000000673 UP000085678 UP000000803 UP000007819 UP000075900 UP000078200 UP000007062 UP000075883 UP000030765 UP000095301 UP000078542 UP000001292 UP000053454 UP000078492 UP000000304 UP000075903 UP000075885 UP000069272 UP000078541 UP000075884 UP000075920 UP000075886 UP000002320 UP000192221 UP000076407 UP000079169 UP000092444 UP000075840 UP000092445 UP000092460 UP000002282 UP000076408 UP000075809 UP000008711 UP000242188 UP000053097 UP000279307 UP000015104 UP000007755 UP000007801 UP000001070 UP000095300 UP000230750 UP000007798 UP000008792 UP000000305 UP000009192 UP000014760
Pfam
Interpro
IPR014729
Rossmann-like_a/b/a_fold
+ More
IPR019407 CTU2
IPR011063 TilS/TtcA_N
IPR036928 AS_sf
IPR023631 Amidase_dom
IPR020556 Amidase_CS
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR016035 Acyl_Trfase/lysoPLipase
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR014043 Acyl_transferase
IPR001227 Ac_transferase_dom_sf
IPR006573 NHR_dom
IPR019407 CTU2
IPR011063 TilS/TtcA_N
IPR036928 AS_sf
IPR023631 Amidase_dom
IPR020556 Amidase_CS
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR016035 Acyl_Trfase/lysoPLipase
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR014043 Acyl_transferase
IPR001227 Ac_transferase_dom_sf
IPR006573 NHR_dom
Gene 3D
CDD
ProteinModelPortal
A0A1E1WG68
A0A2A4JZ41
A0A2H1WWL8
A0A212FIZ1
A0A194RFZ8
A0A0C9RFS3
+ More
U4U4P5 J3JV37 A0A1B6JV90 A0A2P8XGU2 A0A0L7KMA1 D6WYA9 A0A1Y1M3Q4 A0A1B6KH68 A0A067RN66 A0A1B6H2N7 A0A1Y1M405 A0A182H3B6 A0A2K7P687 Q17JB7 A0A293LYM3 A0A158NT32 A0A023GJQ7 V4CK50 T1I792 A0A0P4VUE8 R4FQD1 A0A195BG92 W5JGH8 A0A1S3JE92 A0A0T6AVL9 Q9VIV3 A0A2H8TP24 J9JW98 A0A182RFA1 A0A2R7WS26 A0A1A9V1K9 A0NEF7 A0A182LW56 A0A084W473 A0A1I8MEV7 A0A195C8I0 A0A0V0G776 B4I5W3 A0A0L8HE76 A0A195EFH6 A0A1L8DA21 A0A034VS20 A0A2S2P968 A0A0J9R5L9 B4Q9Z1 A0A182V9Y9 A0A182PHM8 A0A182FHY3 A0A2M4BN88 A0A195EZF8 A0A2M3Z9F5 A0A2R5LK50 A0A182NGW9 A0A182WIH7 A0A023F772 A0A0A1X3V7 A0A182QB04 D3TLU7 B0X911 A0A1W4VER7 A0A182XMW4 A0A3Q0IHQ7 A0A1B0FEC6 A0A1Q3F147 A0A1S3DUI6 A0A182HWS0 A0A1B0A943 A0A1B0C105 B4PAZ6 A0A182Y1A7 A0A2M3Z9D5 A0A151WJD9 B3NM45 A0A210R0K2 A0A026WSU3 T1KDJ5 E9IBE4 F4WG48 B3MN57 A0A2M3ZA98 A0A0K8TW66 B4JCV8 U5EXQ1 A0A0K2T6Y0 A0A1I8NMV7 A0A2G8JTY6 B4MYR2 B4M8J8 E9G223 B4KIW3 R7U248 W8BUL7
U4U4P5 J3JV37 A0A1B6JV90 A0A2P8XGU2 A0A0L7KMA1 D6WYA9 A0A1Y1M3Q4 A0A1B6KH68 A0A067RN66 A0A1B6H2N7 A0A1Y1M405 A0A182H3B6 A0A2K7P687 Q17JB7 A0A293LYM3 A0A158NT32 A0A023GJQ7 V4CK50 T1I792 A0A0P4VUE8 R4FQD1 A0A195BG92 W5JGH8 A0A1S3JE92 A0A0T6AVL9 Q9VIV3 A0A2H8TP24 J9JW98 A0A182RFA1 A0A2R7WS26 A0A1A9V1K9 A0NEF7 A0A182LW56 A0A084W473 A0A1I8MEV7 A0A195C8I0 A0A0V0G776 B4I5W3 A0A0L8HE76 A0A195EFH6 A0A1L8DA21 A0A034VS20 A0A2S2P968 A0A0J9R5L9 B4Q9Z1 A0A182V9Y9 A0A182PHM8 A0A182FHY3 A0A2M4BN88 A0A195EZF8 A0A2M3Z9F5 A0A2R5LK50 A0A182NGW9 A0A182WIH7 A0A023F772 A0A0A1X3V7 A0A182QB04 D3TLU7 B0X911 A0A1W4VER7 A0A182XMW4 A0A3Q0IHQ7 A0A1B0FEC6 A0A1Q3F147 A0A1S3DUI6 A0A182HWS0 A0A1B0A943 A0A1B0C105 B4PAZ6 A0A182Y1A7 A0A2M3Z9D5 A0A151WJD9 B3NM45 A0A210R0K2 A0A026WSU3 T1KDJ5 E9IBE4 F4WG48 B3MN57 A0A2M3ZA98 A0A0K8TW66 B4JCV8 U5EXQ1 A0A0K2T6Y0 A0A1I8NMV7 A0A2G8JTY6 B4MYR2 B4M8J8 E9G223 B4KIW3 R7U248 W8BUL7
Ontologies
GO
PANTHER
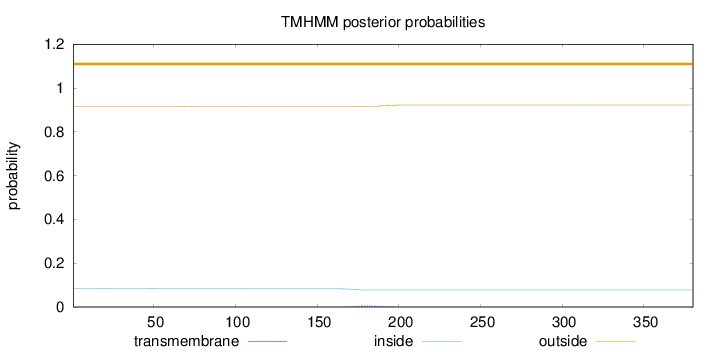
Topology
Subcellular location
Cytoplasm
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14457
Exp number, first 60 AAs:
0.00925
Total prob of N-in:
0.08420
outside
1 - 380
Population Genetic Test Statistics
Pi
190.827782
Theta
208.940535
Tajima's D
-0.265987
CLR
31.858895
CSRT
0.299035048247588
Interpretation
Uncertain
