Gene
KWMTBOMO02730
Pre Gene Modal
BGIBMGA003472
Annotation
PREDICTED:_aryl_hydrocarbon_receptor_nuclear_translocator_homolog_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.074
Sequence
CDS
ATGTCTGCCGTGGCTCCAACTATCCCCGGGGCTGACCCAGCAAAGGATTTACAAAAACGCCGTGCTGGCAGTGTTGGATCAGATGAAGACGACGGAAGTGGTGGAAAATACACAAGGATGGAGGAGGACAATATTCAAGACAAGGAGCGTTTTGCAAGTCGTGAAAATCATTGCGAGATTGAACGTCGCAGGAGGAACAAGATGACGGCGTATATCACGGAACTATCTGATATGGTTCCAACATGCTCCGCACTCGCCCGGAAGCCAGACAAGCTAACCATACTGCGTATGGCAGTAGCTCACATGAAGGCATTGAGAGGCACTGGCAATACTTCTACAGATGGAACATATAAACCATCGTTTTTAACTGATCAAGAATTGAAACATTTAATTTTAGAAGCCGCAGATGGTTTCTTGTTTGTAGTAAGCTGCGATACTGGTCGCATTATTTACGTTAGCGACAGTATAGCACCAGTTCTAAATTATTCACAGGGTGAATGGTACTCGTCGTGTTTATACGACCAAGTGCACCCGGATGACGTTGAAAAAGTACGAGAGCAGTTAAGTACACAAGAACCTCAGAATACGGGTCGTATACTAGACTTGAAGACTGGAACCGTGAAAAAAGAAGGCCACCAATCTTCAATGCGGCAAGTAATGGGTTCACGTCGTGGATTCATCTGTCGGATGCGAGTCGGCGGCACCGCGGAGGGGGCGCACCTCGGCCGGCTGCGAGCGCGCAACTCGCTCGGCGCATCGCACGACGGTCACAACTACGCTGTCGTGCATTGTACAGGATATATCAAAAACTGGCCTCCGACAGGTATGCAAATGGACAGGCCAGTTGAAGATGAACTCCATTCGTCACATTGTTGTCTTGTAGCTATTGGACGATTACAGGTAACTTCGACCCCAAATAATTCAGAAAACAGTTTATGCAATGGTGGAGTTGAATTCGTTTCCCGTCATTCTGTTGACGGCCGCTTCACATTCGCCGATCAGCGCGCAACTCAAGTAATAGGATATGCCCCTGCCGATCTCTTAGGAAAATTGTGTTACGAATTCTATCATCCCGAGGACCAACAACACATGAGAGACAACTTCGATCAAGTATTGAAACTAAAAGGCCAAATAATATCGTTGATGTACCGGTTTCGAACGAAGAGCAGAGAATGGGTTTGGCTGAGAACATCAGCGTTCGCTTTCTTAAATCCGTACAACGACGATGTCGAATATATAGTATGCACCAATACTTTAGCAAATCGGTCGCTGGCGAGTTCAGGCGGTGAGGCAACCCCGGAGGAGGGGTATGACTATCACCTGCGGCACCGTGACGTGTACCCCGCCGCGCCACCCGCTCCCCCTGCGCCCCACGCGCCCGCCGCCCCGTCCGCGCTGCACCAGCATCACCACGCGCCCGCCGCAGAAGTAAGTGGAGTGTATGTGTCAGGCGGGGGAGGGGGTGGGGGCTCGGGCGCGCGCTCCCCCGCGGACACCGGCGCATACCCGGCACACTACGCGCCTAACTACTCGCCGCACCGACCCGCCAACACGCCCCCACACACCACTTGGACCACACTCCGACCGAGCGGCGGTGGCGGCGGCGGTGAGAGCTACGCCTACAGCGGCGAGACCACTGCCGGTGCGCCGCCCGACACCAGTCCAGGACGTTCGCCTCCTGCACCTCCCTATTTACCACCCGCACATTACCACCACAACCATCATCCTCATCCCACGCATGGCATGTGGACATGGCAAGGCGGAGCGGCGGGGGGCGGGGCAGCGGCACCCGAAGGTACGCACGCGCCGCACGAACTCTCGGACATGTTACAGATCCTCGACCAAAGCGGGACTGCAACCTTTGAAGACCTCAACATAAACATGTTTAATTCGAACTTTGAGTAA
Protein
MSAVAPTIPGADPAKDLQKRRAGSVGSDEDDGSGGKYTRMEEDNIQDKERFASRENHCEIERRRRNKMTAYITELSDMVPTCSALARKPDKLTILRMAVAHMKALRGTGNTSTDGTYKPSFLTDQELKHLILEAADGFLFVVSCDTGRIIYVSDSIAPVLNYSQGEWYSSCLYDQVHPDDVEKVREQLSTQEPQNTGRILDLKTGTVKKEGHQSSMRQVMGSRRGFICRMRVGGTAEGAHLGRLRARNSLGASHDGHNYAVVHCTGYIKNWPPTGMQMDRPVEDELHSSHCCLVAIGRLQVTSTPNNSENSLCNGGVEFVSRHSVDGRFTFADQRATQVIGYAPADLLGKLCYEFYHPEDQQHMRDNFDQVLKLKGQIISLMYRFRTKSREWVWLRTSAFAFLNPYNDDVEYIVCTNTLANRSLASSGGEATPEEGYDYHLRHRDVYPAAPPAPPAPHAPAAPSALHQHHHAPAAEVSGVYVSGGGGGGGSGARSPADTGAYPAHYAPNYSPHRPANTPPHTTWTTLRPSGGGGGGESYAYSGETTAGAPPDTSPGRSPPAPPYLPPAHYHHNHHPHPTHGMWTWQGGAAGGGAAAPEGTHAPHELSDMLQILDQSGTATFEDLNINMFNSNFE
Summary
Uniprot
EMBL
BABH01007994
BABH01007995
ODYU01011613
SOQ57468.1
NWSH01000357
PCG77054.1
+ More
PCG77057.1 PCG77056.1 GDQN01008440 JAT82614.1 PCG77055.1 PCG77053.1 PCG77051.1 GAIX01004137 JAA88423.1 AGBW02008321 OWR53694.1 KQ460207 KPJ16735.1 KQ459597 KPI94848.1 JN228345 AFL70632.1 GEZM01007147 JAV95341.1 GEZM01007143 JAV95345.1 KP147913 AKG92750.1 GEDC01002447 JAS34851.1 GEDC01028344 JAS08954.1 KU647692 APU88436.1 KQ981856 KYN34533.1 QOIP01000003 RLU24817.1 GGLE01003378 MBY07504.1
PCG77057.1 PCG77056.1 GDQN01008440 JAT82614.1 PCG77055.1 PCG77053.1 PCG77051.1 GAIX01004137 JAA88423.1 AGBW02008321 OWR53694.1 KQ460207 KPJ16735.1 KQ459597 KPI94848.1 JN228345 AFL70632.1 GEZM01007147 JAV95341.1 GEZM01007143 JAV95345.1 KP147913 AKG92750.1 GEDC01002447 JAS34851.1 GEDC01028344 JAS08954.1 KU647692 APU88436.1 KQ981856 KYN34533.1 QOIP01000003 RLU24817.1 GGLE01003378 MBY07504.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
6E3U
E-value=4.70463e-151,
Score=1373
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
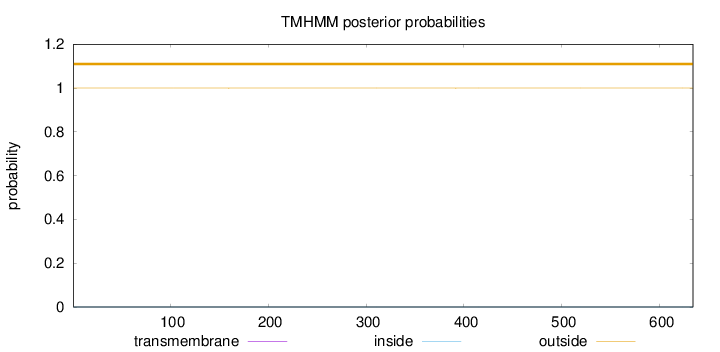
Length:
634
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00274
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00027
outside
1 - 634
Population Genetic Test Statistics
Pi
255.853871
Theta
195.472963
Tajima's D
0.265453
CLR
1.57973
CSRT
0.447627618619069
Interpretation
Uncertain
