Pre Gene Modal
BGIBMGA003470
Annotation
PREDICTED:_tudor_domain-containing_protein_7_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.048
Sequence
CDS
ATGGCGGAAAAAGAACAAGTAATTCAAGCCCTACGTGCAACATTACTATCAGTCAAAGGTGCACTAACTCTGAAACAGTGTAATCGTGATTATAAAGAACTACAAGGAGAATGGATACCATTCAAAAAATTGGGCTATCCGTCATTAGAAAAGCTTTTTTTGGATGTGTCAGGTTTTAAAGTATCCCAAGTTAACGGGGAATGGTATGTAGATGCAATAGCTAGTCAAGAGACCCAACATATTGCTGCTATGGTAGCTCGTCAAAAACCAAGCAAAAAGTCAGGCTCTAAAGTAAATTACTTGAATCGATTACCAAAAAAGCAAGGAATATGGAGGAAGCCAGCTTCAAATAATACATTAAATTATGGAAGTCAGTATTCCAATCAGTACTATAGAGCAAAAAGTGGAGCACCACTTAATAACAACTCTTACAACAACTACAAGAATGTCAAACCTAACAACACGCATAAAACGATCGAAACGAAAAATAATAGTACAATTCCGAAACAGCCAACGAAAACAGCAACGACTGAATACTCAGAAAATACCGCAACGAAAAGTTCTGCTCAGCTTCCACAATTACGAAGTCATAGCTCTGTTTCTGAAGAGCATAAACAATCTGCACGAACAGGCAAAGACATTGAAAATAAAAGTAGCGGAAAAGTTTCTGCGTCACAGAGGCTCTCGAATTTACGAGATCGTCTTAGCACTGTTGATCTGATACCTCTACAATTGCCAGCAGCGAATAATGAAATTGTTCAGGACAAAAGTGACCCGGTAACGAATGTGGTGCCGGCCCATAAATTGGAGGCTCCCCCAGACTGTACGTCCTCAGTAGCGAAACTTGAGTGGACATGTAGAAAGCTAGGCCTCCCATCTCCCGTATACAAAGTGCAGAATATTGGACCGAAACGGGGCCCTGCCAGCTACGACAGTACGGTGAAAGTCGGCGCGTCATACGTCGCGATGTCTTATCCGAATTCTTCGCCTTCCGAAGAACTGGCAAAAGAAGTGGCTGCCGTAAAATTACTTGAACTATTGGAAAGCTCTGAAACTGAAGGAGTACCCACTTCGTCACCAACCAAAGCTATGTCATCTCTCGTGGACCTTGTGTCGGAACATGAAGCTGGACTTTGGGCTAGTACTGTACCTTTTCTTTACAGAAAAAAATATGGCGAAAATTTGCCTACTGACTGGCAAGATCTTGTGGAAAATTGTCCTTTCATCGTGATAGATAGGGTTGCTAACGGACTCTCAGTTCTTCTTCTCGCCAAAGAGGGGTCATTACCATTGCCCGTGGACACGAGTGTGTTCGATGAAGCCATCGAAAGTGACCTGCCTCCGTTACAATTTCCTGAGGACGATTTTTGGAACGTTTTCATCACGGTCGCTAATTCTACGTTAGAAATTTGGTTACGAATAATAGGACCTCAATATAGTGATGCTTTTGAAACACTCCTAGCAGATATGGCAAAATACTACGATAGCTCTGGCACTGCAGTTGACAAATCTATGATTATAAGAGACGCCTGGTATGCGGTCAATTTGGACGACGGCGGCTGGCAACGAGCCAAAATAATGGACTTCGATGAAGAAACAGCCTCCGTATTTTTAGGAGACCACGGAGACGACGACGTCGTGGCTTTAAATAAAATTAGAATTCTAGATTCTAAATTCAGGAAATTACCCCCGCAGGCGATCCTTTGCCGACTGGAAGGCGTGGAGGAGCTGGCGGCGAGCGCGGCGGCGGCGGAGCTGGTGCGGCGCCGACTGCCCGGGGAGGTGCTCGTGGCGGCGCCCGGCGCCCGCACCGACCCCACCGACCCCTCCATCGCGCTCGTGCTCTACGACACCTCCACGCAGCGCGACCTCAACCTCAACAAGGAGATCGTGCACGACTTCTGCATTTCCGGCGCCTTTTCAATCACGCAGAAGCCGTGCGAGGTGGAGGTGAAGTGCGTGACGGAGGAGGGGCGCGTGTGGGTGTCGGCGGCGGGCGGCGCGGACACGGTGCGGGCCGCGCTGGCGCTGCTCACGGCGGGCCCGTACCGCCGCCAGCTGCCCGCCGCGCCCTTCGCGCCCTCGCCCAACGCCGCCACGCTCTACATCGTGCGCACGCTCGCCGGCGACTGGGTTCGCTGCACTATTATAAGCGGCCTGGACGGCGAGGGCACAGTGCGAACGCAACTCGTCGACAGCGGCCTTATCCTGCGGGCTCCCCTGTCATCGTTGGTTCCCTTGCAGACCTTTTCGCCGGCTCTCAACGCCTACCCATTTCAGGCGATGTTGGTCCGCCTGGGCCACGCCGAGCGCGCGGCGGCCGCCATGGTGAGCCGGCTGCGCGAGCTGGTGCTGGACCAGCGCGTGCTGTGCCGAGCGCTCCCCGGCACGGGCGCCGCGTCGTCCGCGCCGCACGTCGAACTGTTCGTCCGCACGGAGCCGCAGAATATGCTCGCCTCCGTCAACAACGACATCTTTATGGAGCACGAGTACCTAAACCAGGACAAGGTGAAAGAGCAGAAGAAGGACACCCCGAATCCCGTAGAGGCGTTAAATAAAAAGAAGGAGCGCTTATACCGGACCGCTAGTGGTGCGGACGAGCGGCCGGAGCGCGCCGCCCTGCCGCCCCCCACCCTGCCCTCCCCCGGACAGTGCTTTGACGTCTACGTGGCGATGGCCGCCAATCCCTGGAATTTTGTGGTTCAACCGAACAGTACGAGACACATGCTACAAACTATGATGTCTTCATTGCAAATTGAATGTCCGAAGCTATCGCAGACAGACGCGCCCCTTAAACCGGCCAATGGGGAGCTGTACACGGTGTATTATGCAAAAGATTTATCGTGGTACAGAGTGACAGTGGCGGGATCTGTGTCGTCGGAGATAGTGTCAGTGTATTTTTGCGACTACGGTGACCTGGCGCTATTCGAAATCCAAGCGCTGCGTCCGGTGCCTCCTACCGTGCCGCTAGCCCGCACGCTGCCCCCGCAGGCCATCAAGGCGCGTCTCTACGACGTATTGCCACTACACCAAGACTGGGCAGTAGAAGATTGCATTAGATTTCAGGAACTGTGCGTAGATCAGCAATACGTAGGTATCTGCAAAGATGTAGGCAAAGATCCATTAAACACAAACGAGCCCTTGTTAACTTTAGAACTTATTGATACGTCCACCGATGAGGATGTTTATTTAAACAAACAATTAGTGTCCGAAGGAAGGGCTAGACTGGCGTCAGCAACCTAG
Protein
MAEKEQVIQALRATLLSVKGALTLKQCNRDYKELQGEWIPFKKLGYPSLEKLFLDVSGFKVSQVNGEWYVDAIASQETQHIAAMVARQKPSKKSGSKVNYLNRLPKKQGIWRKPASNNTLNYGSQYSNQYYRAKSGAPLNNNSYNNYKNVKPNNTHKTIETKNNSTIPKQPTKTATTEYSENTATKSSAQLPQLRSHSSVSEEHKQSARTGKDIENKSSGKVSASQRLSNLRDRLSTVDLIPLQLPAANNEIVQDKSDPVTNVVPAHKLEAPPDCTSSVAKLEWTCRKLGLPSPVYKVQNIGPKRGPASYDSTVKVGASYVAMSYPNSSPSEELAKEVAAVKLLELLESSETEGVPTSSPTKAMSSLVDLVSEHEAGLWASTVPFLYRKKYGENLPTDWQDLVENCPFIVIDRVANGLSVLLLAKEGSLPLPVDTSVFDEAIESDLPPLQFPEDDFWNVFITVANSTLEIWLRIIGPQYSDAFETLLADMAKYYDSSGTAVDKSMIIRDAWYAVNLDDGGWQRAKIMDFDEETASVFLGDHGDDDVVALNKIRILDSKFRKLPPQAILCRLEGVEELAASAAAAELVRRRLPGEVLVAAPGARTDPTDPSIALVLYDTSTQRDLNLNKEIVHDFCISGAFSITQKPCEVEVKCVTEEGRVWVSAAGGADTVRAALALLTAGPYRRQLPAAPFAPSPNAATLYIVRTLAGDWVRCTIISGLDGEGTVRTQLVDSGLILRAPLSSLVPLQTFSPALNAYPFQAMLVRLGHAERAAAAMVSRLRELVLDQRVLCRALPGTGAASSAPHVELFVRTEPQNMLASVNNDIFMEHEYLNQDKVKEQKKDTPNPVEALNKKKERLYRTASGADERPERAALPPPTLPSPGQCFDVYVAMAANPWNFVVQPNSTRHMLQTMMSSLQIECPKLSQTDAPLKPANGELYTVYYAKDLSWYRVTVAGSVSSEIVSVYFCDYGDLALFEIQALRPVPPTVPLARTLPPQAIKARLYDVLPLHQDWAVEDCIRFQELCVDQQYVGICKDVGKDPLNTNEPLLTLELIDTSTDEDVYLNKQLVSEGRARLASAT
Summary
Uniprot
A0A2A4JMJ3
A0A2H1W627
A0A212FIX7
A0A194PUN2
H9J1T3
A0A194RHD0
+ More
A0A088ASD6 K4MU05 A0A2A3E3Q3 A0A0L7R3N2 A0A2P8YNP2 A0A158P239 A0A195FI94 E2B283 A0A151XCS6 F4W6C0 A0A195AVI4 E2BFZ8 A0A151IER4 A0A0J7L633 E9JCT6 A0A1W4WCF0 A0A1W4WCE9 A0A1B6CFC5 A0A1B6CR68 A0A1B6D1B7 A0A0C9RRB8 K7IUE0 A0A293LCK7 A0A2M4BER0 E0W409 A0A2R5L9G3 A0A1B0BX84 A0A182FTK7 A0A1A9XID3 A0A1B0G730 A0A1A9UVJ1 A0A0C9R1B2 A0A0C9Q056 A0A1B0AG76 A0A0K8WBC6 A0A2M4DRT4 W8BDE4 A0A023EWD1 A0A084WLT5 A0A182GEP2 A0A0L0BW13 A0A131Y0R2 A0A0A1X6X8 A0A182QFJ7 A0A147BCJ6 Q0IEJ7 A0A034VCY4 A0A182NSX1 A0A182SY63 A0A0K2UGA7 A0A182KKU4 A0A182M524 A0A182UZ53 A0A182TPJ3 A0A182WDR6 A0A1B6KLK9 A0A195DMK2 A0A182K943 A0A1I8Q3T5 T1PG85 A0A1I8N6Y3 A0A1I8N6Z3 A0A0P4VXZ8 A0A182HW45 A0A1B6JIC1 A0A182RLS6 A0A182WUD1 A0A1B6JIQ0 A0A1B6GV28
A0A088ASD6 K4MU05 A0A2A3E3Q3 A0A0L7R3N2 A0A2P8YNP2 A0A158P239 A0A195FI94 E2B283 A0A151XCS6 F4W6C0 A0A195AVI4 E2BFZ8 A0A151IER4 A0A0J7L633 E9JCT6 A0A1W4WCF0 A0A1W4WCE9 A0A1B6CFC5 A0A1B6CR68 A0A1B6D1B7 A0A0C9RRB8 K7IUE0 A0A293LCK7 A0A2M4BER0 E0W409 A0A2R5L9G3 A0A1B0BX84 A0A182FTK7 A0A1A9XID3 A0A1B0G730 A0A1A9UVJ1 A0A0C9R1B2 A0A0C9Q056 A0A1B0AG76 A0A0K8WBC6 A0A2M4DRT4 W8BDE4 A0A023EWD1 A0A084WLT5 A0A182GEP2 A0A0L0BW13 A0A131Y0R2 A0A0A1X6X8 A0A182QFJ7 A0A147BCJ6 Q0IEJ7 A0A034VCY4 A0A182NSX1 A0A182SY63 A0A0K2UGA7 A0A182KKU4 A0A182M524 A0A182UZ53 A0A182TPJ3 A0A182WDR6 A0A1B6KLK9 A0A195DMK2 A0A182K943 A0A1I8Q3T5 T1PG85 A0A1I8N6Y3 A0A1I8N6Z3 A0A0P4VXZ8 A0A182HW45 A0A1B6JIC1 A0A182RLS6 A0A182WUD1 A0A1B6JIQ0 A0A1B6GV28
Pubmed
EMBL
NWSH01000990
PCG73201.1
ODYU01006578
SOQ48541.1
AGBW02008321
OWR53698.1
+ More
KQ459597 KPI94845.1 BABH01008001 BABH01008002 BABH01008003 BABH01008004 KQ460207 KPJ16730.1 JQ434104 AFV31612.1 KZ288425 PBC25922.1 KQ414663 KOC65492.1 PYGN01000464 PSN45877.1 ADTU01006976 KQ981523 KYN40395.1 GL445067 EFN60209.1 KQ982297 KYQ58172.1 GL887707 EGI70255.1 KQ976731 KYM76243.1 GL448096 EFN85400.1 KQ977858 KYM99296.1 LBMM01000553 KMQ98076.1 GL771864 EFZ09369.1 GEDC01027454 GEDC01025258 JAS09844.1 JAS12040.1 GEDC01029177 GEDC01021390 JAS08121.1 JAS15908.1 GEDC01017824 GEDC01006540 JAS19474.1 JAS30758.1 GBYB01009866 JAG79633.1 AAZX01006627 GFWV01000831 MAA25561.1 GGFJ01002300 MBW51441.1 DS235886 EEB20365.1 GGLE01002015 MBY06141.1 JXJN01022114 CCAG010003770 GBYB01001745 JAG71512.1 GBYB01007218 JAG76985.1 GDHF01007845 GDHF01003975 JAI44469.1 JAI48339.1 GGFL01016107 MBW80285.1 GAMC01009843 JAB96712.1 GAPW01000048 JAC13550.1 ATLV01024278 KE525351 KFB51179.1 JXUM01011731 JXUM01011732 KQ560337 KXJ82979.1 JRES01001352 KNC23404.1 GEFM01004210 JAP71586.1 GBXI01007792 JAD06500.1 AXCN02002134 AXCN02002135 GEGO01006911 JAR88493.1 CH477655 EAT37713.1 GAKP01018975 JAC39977.1 HACA01019621 CDW36982.1 AXCM01004550 AXCM01004551 GEBQ01027625 JAT12352.1 KQ980724 KYN14061.1 KA647822 AFP62451.1 GDRN01099291 JAI58798.1 APCN01001462 GECU01009043 JAS98663.1 GECU01008555 JAS99151.1 GECZ01003465 JAS66304.1
KQ459597 KPI94845.1 BABH01008001 BABH01008002 BABH01008003 BABH01008004 KQ460207 KPJ16730.1 JQ434104 AFV31612.1 KZ288425 PBC25922.1 KQ414663 KOC65492.1 PYGN01000464 PSN45877.1 ADTU01006976 KQ981523 KYN40395.1 GL445067 EFN60209.1 KQ982297 KYQ58172.1 GL887707 EGI70255.1 KQ976731 KYM76243.1 GL448096 EFN85400.1 KQ977858 KYM99296.1 LBMM01000553 KMQ98076.1 GL771864 EFZ09369.1 GEDC01027454 GEDC01025258 JAS09844.1 JAS12040.1 GEDC01029177 GEDC01021390 JAS08121.1 JAS15908.1 GEDC01017824 GEDC01006540 JAS19474.1 JAS30758.1 GBYB01009866 JAG79633.1 AAZX01006627 GFWV01000831 MAA25561.1 GGFJ01002300 MBW51441.1 DS235886 EEB20365.1 GGLE01002015 MBY06141.1 JXJN01022114 CCAG010003770 GBYB01001745 JAG71512.1 GBYB01007218 JAG76985.1 GDHF01007845 GDHF01003975 JAI44469.1 JAI48339.1 GGFL01016107 MBW80285.1 GAMC01009843 JAB96712.1 GAPW01000048 JAC13550.1 ATLV01024278 KE525351 KFB51179.1 JXUM01011731 JXUM01011732 KQ560337 KXJ82979.1 JRES01001352 KNC23404.1 GEFM01004210 JAP71586.1 GBXI01007792 JAD06500.1 AXCN02002134 AXCN02002135 GEGO01006911 JAR88493.1 CH477655 EAT37713.1 GAKP01018975 JAC39977.1 HACA01019621 CDW36982.1 AXCM01004550 AXCM01004551 GEBQ01027625 JAT12352.1 KQ980724 KYN14061.1 KA647822 AFP62451.1 GDRN01099291 JAI58798.1 APCN01001462 GECU01009043 JAS98663.1 GECU01008555 JAS99151.1 GECZ01003465 JAS66304.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000005204
UP000053240
UP000005203
+ More
UP000242457 UP000053825 UP000245037 UP000005205 UP000078541 UP000000311 UP000075809 UP000007755 UP000078540 UP000008237 UP000078542 UP000036403 UP000192223 UP000002358 UP000009046 UP000092460 UP000069272 UP000092443 UP000092444 UP000078200 UP000092445 UP000030765 UP000069940 UP000249989 UP000037069 UP000075886 UP000008820 UP000075884 UP000075901 UP000075882 UP000075883 UP000075903 UP000075902 UP000075920 UP000078492 UP000075881 UP000095300 UP000095301 UP000075840 UP000075900 UP000076407
UP000242457 UP000053825 UP000245037 UP000005205 UP000078541 UP000000311 UP000075809 UP000007755 UP000078540 UP000008237 UP000078542 UP000036403 UP000192223 UP000002358 UP000009046 UP000092460 UP000069272 UP000092443 UP000092444 UP000078200 UP000092445 UP000030765 UP000069940 UP000249989 UP000037069 UP000075886 UP000008820 UP000075884 UP000075901 UP000075882 UP000075883 UP000075903 UP000075902 UP000075920 UP000078492 UP000075881 UP000095300 UP000095301 UP000075840 UP000075900 UP000076407
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JMJ3
A0A2H1W627
A0A212FIX7
A0A194PUN2
H9J1T3
A0A194RHD0
+ More
A0A088ASD6 K4MU05 A0A2A3E3Q3 A0A0L7R3N2 A0A2P8YNP2 A0A158P239 A0A195FI94 E2B283 A0A151XCS6 F4W6C0 A0A195AVI4 E2BFZ8 A0A151IER4 A0A0J7L633 E9JCT6 A0A1W4WCF0 A0A1W4WCE9 A0A1B6CFC5 A0A1B6CR68 A0A1B6D1B7 A0A0C9RRB8 K7IUE0 A0A293LCK7 A0A2M4BER0 E0W409 A0A2R5L9G3 A0A1B0BX84 A0A182FTK7 A0A1A9XID3 A0A1B0G730 A0A1A9UVJ1 A0A0C9R1B2 A0A0C9Q056 A0A1B0AG76 A0A0K8WBC6 A0A2M4DRT4 W8BDE4 A0A023EWD1 A0A084WLT5 A0A182GEP2 A0A0L0BW13 A0A131Y0R2 A0A0A1X6X8 A0A182QFJ7 A0A147BCJ6 Q0IEJ7 A0A034VCY4 A0A182NSX1 A0A182SY63 A0A0K2UGA7 A0A182KKU4 A0A182M524 A0A182UZ53 A0A182TPJ3 A0A182WDR6 A0A1B6KLK9 A0A195DMK2 A0A182K943 A0A1I8Q3T5 T1PG85 A0A1I8N6Y3 A0A1I8N6Z3 A0A0P4VXZ8 A0A182HW45 A0A1B6JIC1 A0A182RLS6 A0A182WUD1 A0A1B6JIQ0 A0A1B6GV28
A0A088ASD6 K4MU05 A0A2A3E3Q3 A0A0L7R3N2 A0A2P8YNP2 A0A158P239 A0A195FI94 E2B283 A0A151XCS6 F4W6C0 A0A195AVI4 E2BFZ8 A0A151IER4 A0A0J7L633 E9JCT6 A0A1W4WCF0 A0A1W4WCE9 A0A1B6CFC5 A0A1B6CR68 A0A1B6D1B7 A0A0C9RRB8 K7IUE0 A0A293LCK7 A0A2M4BER0 E0W409 A0A2R5L9G3 A0A1B0BX84 A0A182FTK7 A0A1A9XID3 A0A1B0G730 A0A1A9UVJ1 A0A0C9R1B2 A0A0C9Q056 A0A1B0AG76 A0A0K8WBC6 A0A2M4DRT4 W8BDE4 A0A023EWD1 A0A084WLT5 A0A182GEP2 A0A0L0BW13 A0A131Y0R2 A0A0A1X6X8 A0A182QFJ7 A0A147BCJ6 Q0IEJ7 A0A034VCY4 A0A182NSX1 A0A182SY63 A0A0K2UGA7 A0A182KKU4 A0A182M524 A0A182UZ53 A0A182TPJ3 A0A182WDR6 A0A1B6KLK9 A0A195DMK2 A0A182K943 A0A1I8Q3T5 T1PG85 A0A1I8N6Y3 A0A1I8N6Z3 A0A0P4VXZ8 A0A182HW45 A0A1B6JIC1 A0A182RLS6 A0A182WUD1 A0A1B6JIQ0 A0A1B6GV28
PDB
5M9N
E-value=6.57543e-08,
Score=140
Ontologies
GO
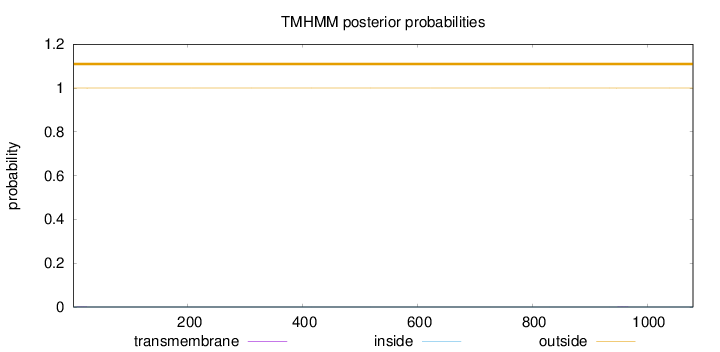
Topology
Length:
1080
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01021
Exp number, first 60 AAs:
0.00414
Total prob of N-in:
0.00025
outside
1 - 1080
Population Genetic Test Statistics
Pi
212.006572
Theta
197.973553
Tajima's D
0.739697
CLR
0.650249
CSRT
0.587770611469427
Interpretation
Uncertain
