Gene
KWMTBOMO02726 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003725
Annotation
PREDICTED:_glycylpeptide_N-tetradecanoyltransferase_2_[Amyelois_transitella]
Full name
Glycylpeptide N-tetradecanoyltransferase
Location in the cell
Cytoplasmic Reliability : 3.078
Sequence
CDS
ATGGAAGAAACGAAAACGGGCGTATCTTGTGATTTTAGTAAAGAAAACGATGAGAAACCTCAGAAAAAGAAAAATAAAAACAAGAAAAAACGTGGTGGAGACGGAGATCATGGTATATCTACGGCAAATGAGAGTTCGTCATCAATGCCAGATCCAACTGAAGGTCATCAAAACATTTCATTAAAAGACTTAAAAATGGCAATGGAGGTACTAAATTTACAGCAAAAGCCTGCAAAAACTACAGAGGAGGCATTGCATAAAGCATACCAATTCTGGTCAACCCAACCGGTGCCAAAAATGGATGAAAAAATTATTGCCAACGAGCCTATAGAACTACCAAAATCACCGGAAGAAATAAGGGCAGAACCCTATACTTTACCTGACGGATTCCAATGGGATACTTTGAATTTGAACGAACCTTTAGTATTGAAAGAACTGTACACTTTACTTAATGAAAATTATGTGGAAGATGATGATTGTATGTTTCGTTTTGATTACCAAACTGATTTTTTGAAATGGGCTCTACAACCACCTGGTTGGAAAATGGAATGGCATTGTGGTGTTCGTGTGGTGAAATCTGGACGGCTTGTTGGTTTCATTTCAGCAACTCCTGCTGATTTAAGAATATATAACCATGTTCAAACTGTTGTTGAAATAAATTTTCTTTGTGTGCACAAAAAATTGCGTTCTAAGCGAGTAGCTCCTGTCTTAATAAGGGAAATAACAAGAAGAGTTAACTTAACTGGTATATTCCAGGGTGTGTATACAGCTGGAATTGTGTTGCCTAAGCCTATTGCCACATGTCGCTATTGGCATAGGTCTCTTAATCCAAAGAAATTAATTGACATAAAATTCAGTCATTTGTCACGTAATATGACAATGCAAAGAACGCTTAAACTCTTTAAGTTACCAGACATGCCAAAAACACCAGGTTTCCGCAAAATGGAGATTCAAGACTGTGAAGCAGTAGTAAAATTACTCAATGATTACTTAAAAAAATTTGATTTGGCACCTATATTTTCTGATGAAGATTTTAAACATTGGTTTACACCACAAGTGGGAATAATTGATAGTTTTGTTGTGGAAGGTCCTAATTCTACAATAACTGATTTCGTCAGTTATTATACATTGCCTTCAACTGTTGTTTACCATCCAGTGCACAAAACACTAAAAGCAGCTTATTCATTCTATAATGTGTCAACCAAGACACCTTGGGTGGATTTGATGCTGGATGCATTAATAACAGCAAAAAATTCAGGATTTGATGTGTTTAATGCACTAGATTTGATGGATAACAAAGAATTTCTTGAACCTCTGAAATTTGGGATTGGTGATGGAAATTTGCAATATTATTTGTATAATTGGAGATGTCCTAGTATGGCCCCGAATAAAATTGGTCTGGTACTTCAATAA
Protein
MEETKTGVSCDFSKENDEKPQKKKNKNKKKRGGDGDHGISTANESSSSMPDPTEGHQNISLKDLKMAMEVLNLQQKPAKTTEEALHKAYQFWSTQPVPKMDEKIIANEPIELPKSPEEIRAEPYTLPDGFQWDTLNLNEPLVLKELYTLLNENYVEDDDCMFRFDYQTDFLKWALQPPGWKMEWHCGVRVVKSGRLVGFISATPADLRIYNHVQTVVEINFLCVHKKLRSKRVAPVLIREITRRVNLTGIFQGVYTAGIVLPKPIATCRYWHRSLNPKKLIDIKFSHLSRNMTMQRTLKLFKLPDMPKTPGFRKMEIQDCEAVVKLLNDYLKKFDLAPIFSDEDFKHWFTPQVGIIDSFVVEGPNSTITDFVSYYTLPSTVVYHPVHKTLKAAYSFYNVSTKTPWVDLMLDALITAKNSGFDVFNALDLMDNKEFLEPLKFGIGDGNLQYYLYNWRCPSMAPNKIGLVLQ
Summary
Description
Adds a myristoyl group to the N-terminal glycine residue of certain cellular proteins.
Catalytic Activity
N-terminal glycyl-[protein] + tetradecanoyl-CoA = CoA + H(+) + N-tetradecanoylglycyl-[protein]
Similarity
Belongs to the NMT family.
Uniprot
H9J2I8
A0A2H1WVX7
A0A2W1B971
A0A1E1W6D7
A0A2A4JN47
A0A194PPD7
+ More
A0A194RFY9 A0A0L7KPX0 A0A212FJ00 S4NK99 A0A1Y1N3L6 A0A1W4WFE4 A0A1B6DI98 A0A2J7QUZ0 A0A0C9QK64 A0A067QR46 A0A0T6BBQ2 E9IQ13 A0A151I280 A0A151J669 F4WSI5 A0A158NRE0 A0A0A9X9A8 A0A224XPY2 A0A0V0G975 A0A151X618 A0A026X086 A0A195EXL0 S6D9I6 A0A069DU61 A0A154P1P5 A0A1B6M6A3 E2ADC0 K7INE9 A0JCZ1 E2C0S6 A0A0L7RCQ8 B7S8A8 A0A0M9AB74 A0A088AMF3 V5G9K7 T1HP74 A0A2A3EHH8 A0A0A9XA49 D2A651 U5EYD7 U4UGU2 U4U9S9 A0A182HGS8 A0A084VAJ6 Q7PY97 A0A182X2Q9 A0A182KMB6 A0A2H8TF45 A0A182JPZ5 A0A182QCW1 A0A182P9S8 A0A182JGT2 A0A182XV91 A0A2S2QCQ3 U5EP13 A0A182F6U9 A0A2M4ADW8 A0A182VA87 A0A182M789 A0A2M4BIG1 A0A182VRI7 A0A2M4ACJ1 A0A2M4BIG0 A0A2M4BJ70 A0A182RNZ5 W5JKM4 A0A182UJK6 A0A182NFK0 A0A1E1XGS0 A0A023FMG3 A0A131XG87 A0A2M3ZGY0 L7LXT0 A0A224YQS2 A0A131Z756 J9JVX2 B0XIR1 T1E203 A0A2M3Z7U4 A0A1Q3F6P2 N6T6Q1 Q17B42 A0A0P6ATE7 A0A0P4ZE44 A0A182H5P0 A0A0P5DMZ0 A0A0P5UAJ8 A0A0N8CL07 A0A0N7ZSE4 A0A0N7ZNT8 A0A0P6JA25 A0A0P5CD89 A0A0N7ZPP4 A0A0P4ZMQ1
A0A194RFY9 A0A0L7KPX0 A0A212FJ00 S4NK99 A0A1Y1N3L6 A0A1W4WFE4 A0A1B6DI98 A0A2J7QUZ0 A0A0C9QK64 A0A067QR46 A0A0T6BBQ2 E9IQ13 A0A151I280 A0A151J669 F4WSI5 A0A158NRE0 A0A0A9X9A8 A0A224XPY2 A0A0V0G975 A0A151X618 A0A026X086 A0A195EXL0 S6D9I6 A0A069DU61 A0A154P1P5 A0A1B6M6A3 E2ADC0 K7INE9 A0JCZ1 E2C0S6 A0A0L7RCQ8 B7S8A8 A0A0M9AB74 A0A088AMF3 V5G9K7 T1HP74 A0A2A3EHH8 A0A0A9XA49 D2A651 U5EYD7 U4UGU2 U4U9S9 A0A182HGS8 A0A084VAJ6 Q7PY97 A0A182X2Q9 A0A182KMB6 A0A2H8TF45 A0A182JPZ5 A0A182QCW1 A0A182P9S8 A0A182JGT2 A0A182XV91 A0A2S2QCQ3 U5EP13 A0A182F6U9 A0A2M4ADW8 A0A182VA87 A0A182M789 A0A2M4BIG1 A0A182VRI7 A0A2M4ACJ1 A0A2M4BIG0 A0A2M4BJ70 A0A182RNZ5 W5JKM4 A0A182UJK6 A0A182NFK0 A0A1E1XGS0 A0A023FMG3 A0A131XG87 A0A2M3ZGY0 L7LXT0 A0A224YQS2 A0A131Z756 J9JVX2 B0XIR1 T1E203 A0A2M3Z7U4 A0A1Q3F6P2 N6T6Q1 Q17B42 A0A0P6ATE7 A0A0P4ZE44 A0A182H5P0 A0A0P5DMZ0 A0A0P5UAJ8 A0A0N8CL07 A0A0N7ZSE4 A0A0N7ZNT8 A0A0P6JA25 A0A0P5CD89 A0A0N7ZPP4 A0A0P4ZMQ1
EC Number
2.3.1.97
Pubmed
19121390
28756777
26354079
26227816
22118469
23622113
+ More
28004739 24845553 21282665 21719571 21347285 25401762 26823975 24508170 30249741 23938757 26334808 20798317 20075255 18362917 19820115 23537049 24438588 12364791 14747013 17210077 20966253 25244985 20920257 23761445 28503490 28049606 25576852 28797301 26830274 24330624 17510324 26483478
28004739 24845553 21282665 21719571 21347285 25401762 26823975 24508170 30249741 23938757 26334808 20798317 20075255 18362917 19820115 23537049 24438588 12364791 14747013 17210077 20966253 25244985 20920257 23761445 28503490 28049606 25576852 28797301 26830274 24330624 17510324 26483478
EMBL
BABH01008004
ODYU01011465
SOQ57210.1
KZ150495
PZC70674.1
GDQN01008502
+ More
JAT82552.1 NWSH01000990 PCG73206.1 KQ459597 KPI94843.1 KQ460207 KPJ16728.1 JTDY01007562 KOB65111.1 AGBW02008321 OWR53700.1 GAIX01013389 JAA79171.1 GEZM01013523 JAV92511.1 GEDC01011943 JAS25355.1 NEVH01010480 PNF32398.1 GBYB01000997 GBYB01000998 JAG70764.1 JAG70765.1 KK853123 KDR11094.1 LJIG01002182 KRT84758.1 GL764659 EFZ17329.1 KQ976549 KYM80904.1 KQ979928 KYN18602.1 GL888322 EGI62850.1 ADTU01023942 GBHO01027391 GBHO01027388 GBRD01013269 GDHC01000425 JAG16213.1 JAG16216.1 JAG52557.1 JAQ18204.1 GFTR01006355 JAW10071.1 GECL01001663 JAP04461.1 KQ982494 KYQ55740.1 KK107054 QOIP01000004 EZA61503.1 RLU24128.1 KQ981923 KYN32963.1 HF586472 CCQ71106.1 GBGD01001429 JAC87460.1 KQ434796 KZC05777.1 GEBQ01008539 JAT31438.1 GL438749 EFN68536.1 AC191960 ABK57056.1 GL451846 EFN78470.1 KQ414615 KOC68536.1 EF710644 ACE75132.1 KQ435711 KOX79589.1 GALX01001641 JAB66825.1 ACPB03012986 KZ288248 PBC31158.1 GBHO01027941 GBHO01027392 GBRD01013271 JAG15663.1 JAG16212.1 JAG52555.1 KQ971346 EFA05466.1 GANO01000362 JAB59509.1 KB632191 ERL89816.1 ERL89817.1 APCN01005945 ATLV01004127 KE524190 KFB34990.1 AAAB01008987 EAA01164.5 GFXV01000896 MBW12701.1 AXCN02000208 GGMS01006304 MBY75507.1 GANO01000361 JAB59510.1 GGFK01005663 MBW38984.1 AXCM01002852 GGFJ01003698 MBW52839.1 GGFK01005121 MBW38442.1 GGFJ01003699 MBW52840.1 GGFJ01003697 MBW52838.1 ADMH02001255 ETN63435.1 GFAC01000701 JAT98487.1 GBBK01002442 JAC22040.1 GEFH01002067 JAP66514.1 GGFM01007018 MBW27769.1 GACK01008632 JAA56402.1 GFPF01008119 MAA19265.1 GEDV01002291 JAP86266.1 ABLF02023070 DS233355 EDS29583.1 GALA01001189 JAA93663.1 GGFM01003824 MBW24575.1 GFDL01011897 JAV23148.1 APGK01050185 APGK01050186 KB741167 ENN73373.1 CH477326 EAT43469.1 GDIP01024709 JAM79006.1 GDIP01227232 GDIP01135157 LRGB01001151 JAI96169.1 KZS13435.1 JXUM01112158 KQ565567 KXJ70872.1 GDIP01154093 JAJ69309.1 GDIP01119452 JAL84262.1 GDIP01121391 JAL82323.1 GDIP01217185 JAJ06217.1 GDIP01227233 JAI96168.1 GDIQ01010513 JAN84224.1 GDIP01188229 JAJ35173.1 GDIP01224785 JAI98616.1 GDIP01214779 JAJ08623.1
JAT82552.1 NWSH01000990 PCG73206.1 KQ459597 KPI94843.1 KQ460207 KPJ16728.1 JTDY01007562 KOB65111.1 AGBW02008321 OWR53700.1 GAIX01013389 JAA79171.1 GEZM01013523 JAV92511.1 GEDC01011943 JAS25355.1 NEVH01010480 PNF32398.1 GBYB01000997 GBYB01000998 JAG70764.1 JAG70765.1 KK853123 KDR11094.1 LJIG01002182 KRT84758.1 GL764659 EFZ17329.1 KQ976549 KYM80904.1 KQ979928 KYN18602.1 GL888322 EGI62850.1 ADTU01023942 GBHO01027391 GBHO01027388 GBRD01013269 GDHC01000425 JAG16213.1 JAG16216.1 JAG52557.1 JAQ18204.1 GFTR01006355 JAW10071.1 GECL01001663 JAP04461.1 KQ982494 KYQ55740.1 KK107054 QOIP01000004 EZA61503.1 RLU24128.1 KQ981923 KYN32963.1 HF586472 CCQ71106.1 GBGD01001429 JAC87460.1 KQ434796 KZC05777.1 GEBQ01008539 JAT31438.1 GL438749 EFN68536.1 AC191960 ABK57056.1 GL451846 EFN78470.1 KQ414615 KOC68536.1 EF710644 ACE75132.1 KQ435711 KOX79589.1 GALX01001641 JAB66825.1 ACPB03012986 KZ288248 PBC31158.1 GBHO01027941 GBHO01027392 GBRD01013271 JAG15663.1 JAG16212.1 JAG52555.1 KQ971346 EFA05466.1 GANO01000362 JAB59509.1 KB632191 ERL89816.1 ERL89817.1 APCN01005945 ATLV01004127 KE524190 KFB34990.1 AAAB01008987 EAA01164.5 GFXV01000896 MBW12701.1 AXCN02000208 GGMS01006304 MBY75507.1 GANO01000361 JAB59510.1 GGFK01005663 MBW38984.1 AXCM01002852 GGFJ01003698 MBW52839.1 GGFK01005121 MBW38442.1 GGFJ01003699 MBW52840.1 GGFJ01003697 MBW52838.1 ADMH02001255 ETN63435.1 GFAC01000701 JAT98487.1 GBBK01002442 JAC22040.1 GEFH01002067 JAP66514.1 GGFM01007018 MBW27769.1 GACK01008632 JAA56402.1 GFPF01008119 MAA19265.1 GEDV01002291 JAP86266.1 ABLF02023070 DS233355 EDS29583.1 GALA01001189 JAA93663.1 GGFM01003824 MBW24575.1 GFDL01011897 JAV23148.1 APGK01050185 APGK01050186 KB741167 ENN73373.1 CH477326 EAT43469.1 GDIP01024709 JAM79006.1 GDIP01227232 GDIP01135157 LRGB01001151 JAI96169.1 KZS13435.1 JXUM01112158 KQ565567 KXJ70872.1 GDIP01154093 JAJ69309.1 GDIP01119452 JAL84262.1 GDIP01121391 JAL82323.1 GDIP01217185 JAJ06217.1 GDIP01227233 JAI96168.1 GDIQ01010513 JAN84224.1 GDIP01188229 JAJ35173.1 GDIP01224785 JAI98616.1 GDIP01214779 JAJ08623.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000192223 UP000235965 UP000027135 UP000078540 UP000078492 UP000007755 UP000005205 UP000075809 UP000053097 UP000279307 UP000078541 UP000076502 UP000000311 UP000002358 UP000008237 UP000053825 UP000053105 UP000005203 UP000015103 UP000242457 UP000007266 UP000030742 UP000075840 UP000030765 UP000007062 UP000076407 UP000075882 UP000075881 UP000075886 UP000075885 UP000075880 UP000076408 UP000069272 UP000075903 UP000075883 UP000075920 UP000075900 UP000000673 UP000075902 UP000075884 UP000007819 UP000002320 UP000019118 UP000008820 UP000076858 UP000069940 UP000249989
UP000192223 UP000235965 UP000027135 UP000078540 UP000078492 UP000007755 UP000005205 UP000075809 UP000053097 UP000279307 UP000078541 UP000076502 UP000000311 UP000002358 UP000008237 UP000053825 UP000053105 UP000005203 UP000015103 UP000242457 UP000007266 UP000030742 UP000075840 UP000030765 UP000007062 UP000076407 UP000075882 UP000075881 UP000075886 UP000075885 UP000075880 UP000076408 UP000069272 UP000075903 UP000075883 UP000075920 UP000075900 UP000000673 UP000075902 UP000075884 UP000007819 UP000002320 UP000019118 UP000008820 UP000076858 UP000069940 UP000249989
Interpro
IPR016181
Acyl_CoA_acyltransferase
+ More
IPR022678 MyristoylCoA_TrFase_CS
IPR022676 MyristoylCoA_TrFase_N
IPR022677 MyristoylCoA_TrFase_C
IPR000903 MyristoylCoA_TrFase
IPR027417 P-loop_NTPase
IPR017907 Znf_RING_CS
IPR034085 TOG
IPR038737 SF3b_su1-like
IPR011989 ARM-like
IPR015016 SF3b_su1
IPR001841 Znf_RING
IPR001806 Small_GTPase
IPR032443 RAWUL
IPR005225 Small_GTP-bd_dom
IPR016024 ARM-type_fold
IPR013083 Znf_RING/FYVE/PHD
IPR022678 MyristoylCoA_TrFase_CS
IPR022676 MyristoylCoA_TrFase_N
IPR022677 MyristoylCoA_TrFase_C
IPR000903 MyristoylCoA_TrFase
IPR027417 P-loop_NTPase
IPR017907 Znf_RING_CS
IPR034085 TOG
IPR038737 SF3b_su1-like
IPR011989 ARM-like
IPR015016 SF3b_su1
IPR001841 Znf_RING
IPR001806 Small_GTPase
IPR032443 RAWUL
IPR005225 Small_GTP-bd_dom
IPR016024 ARM-type_fold
IPR013083 Znf_RING/FYVE/PHD
Gene 3D
ProteinModelPortal
H9J2I8
A0A2H1WVX7
A0A2W1B971
A0A1E1W6D7
A0A2A4JN47
A0A194PPD7
+ More
A0A194RFY9 A0A0L7KPX0 A0A212FJ00 S4NK99 A0A1Y1N3L6 A0A1W4WFE4 A0A1B6DI98 A0A2J7QUZ0 A0A0C9QK64 A0A067QR46 A0A0T6BBQ2 E9IQ13 A0A151I280 A0A151J669 F4WSI5 A0A158NRE0 A0A0A9X9A8 A0A224XPY2 A0A0V0G975 A0A151X618 A0A026X086 A0A195EXL0 S6D9I6 A0A069DU61 A0A154P1P5 A0A1B6M6A3 E2ADC0 K7INE9 A0JCZ1 E2C0S6 A0A0L7RCQ8 B7S8A8 A0A0M9AB74 A0A088AMF3 V5G9K7 T1HP74 A0A2A3EHH8 A0A0A9XA49 D2A651 U5EYD7 U4UGU2 U4U9S9 A0A182HGS8 A0A084VAJ6 Q7PY97 A0A182X2Q9 A0A182KMB6 A0A2H8TF45 A0A182JPZ5 A0A182QCW1 A0A182P9S8 A0A182JGT2 A0A182XV91 A0A2S2QCQ3 U5EP13 A0A182F6U9 A0A2M4ADW8 A0A182VA87 A0A182M789 A0A2M4BIG1 A0A182VRI7 A0A2M4ACJ1 A0A2M4BIG0 A0A2M4BJ70 A0A182RNZ5 W5JKM4 A0A182UJK6 A0A182NFK0 A0A1E1XGS0 A0A023FMG3 A0A131XG87 A0A2M3ZGY0 L7LXT0 A0A224YQS2 A0A131Z756 J9JVX2 B0XIR1 T1E203 A0A2M3Z7U4 A0A1Q3F6P2 N6T6Q1 Q17B42 A0A0P6ATE7 A0A0P4ZE44 A0A182H5P0 A0A0P5DMZ0 A0A0P5UAJ8 A0A0N8CL07 A0A0N7ZSE4 A0A0N7ZNT8 A0A0P6JA25 A0A0P5CD89 A0A0N7ZPP4 A0A0P4ZMQ1
A0A194RFY9 A0A0L7KPX0 A0A212FJ00 S4NK99 A0A1Y1N3L6 A0A1W4WFE4 A0A1B6DI98 A0A2J7QUZ0 A0A0C9QK64 A0A067QR46 A0A0T6BBQ2 E9IQ13 A0A151I280 A0A151J669 F4WSI5 A0A158NRE0 A0A0A9X9A8 A0A224XPY2 A0A0V0G975 A0A151X618 A0A026X086 A0A195EXL0 S6D9I6 A0A069DU61 A0A154P1P5 A0A1B6M6A3 E2ADC0 K7INE9 A0JCZ1 E2C0S6 A0A0L7RCQ8 B7S8A8 A0A0M9AB74 A0A088AMF3 V5G9K7 T1HP74 A0A2A3EHH8 A0A0A9XA49 D2A651 U5EYD7 U4UGU2 U4U9S9 A0A182HGS8 A0A084VAJ6 Q7PY97 A0A182X2Q9 A0A182KMB6 A0A2H8TF45 A0A182JPZ5 A0A182QCW1 A0A182P9S8 A0A182JGT2 A0A182XV91 A0A2S2QCQ3 U5EP13 A0A182F6U9 A0A2M4ADW8 A0A182VA87 A0A182M789 A0A2M4BIG1 A0A182VRI7 A0A2M4ACJ1 A0A2M4BIG0 A0A2M4BJ70 A0A182RNZ5 W5JKM4 A0A182UJK6 A0A182NFK0 A0A1E1XGS0 A0A023FMG3 A0A131XG87 A0A2M3ZGY0 L7LXT0 A0A224YQS2 A0A131Z756 J9JVX2 B0XIR1 T1E203 A0A2M3Z7U4 A0A1Q3F6P2 N6T6Q1 Q17B42 A0A0P6ATE7 A0A0P4ZE44 A0A182H5P0 A0A0P5DMZ0 A0A0P5UAJ8 A0A0N8CL07 A0A0N7ZSE4 A0A0N7ZNT8 A0A0P6JA25 A0A0P5CD89 A0A0N7ZPP4 A0A0P4ZMQ1
PDB
1RXT
E-value=3.79993e-168,
Score=1519
Ontologies
GO
Topology
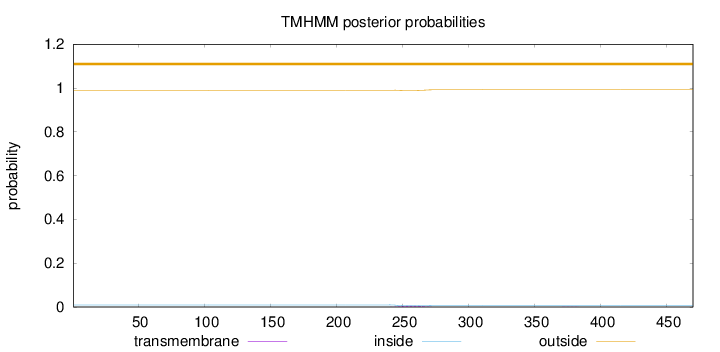
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10551
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00952
outside
1 - 470
Population Genetic Test Statistics
Pi
242.783172
Theta
219.178584
Tajima's D
0.339107
CLR
0.157768
CSRT
0.468976551172441
Interpretation
Uncertain
