Gene
KWMTBOMO02722
Pre Gene Modal
BGIBMGA003727
Annotation
PREDICTED:_exostosin-1_isoform_X2_[Papilio_machaon]
Full name
Exostosin-1
Alternative Name
Protein tout-velu
Location in the cell
PlasmaMembrane Reliability : 1.937
Sequence
CDS
ATGGATACATGTTTCGACTTTTCTAAATGTGGAAGTGATCCAAAAATTTTTGTGTACCCAAGTGATGGGACTGTCAGTGCTTCATATAGGAAGGTATTATCAGTAATAAGAGAGTCCAGATATGTAACTAGAGATCCAAATGAAGCATGTTTATTTGTGCCAGCTATTGATACACTGGATGCTGACCCCTTGTCATCAGAACATGTATCTGATGTCGCTTCACGTCTATCGAGATTACCTTATTGGAGAAATGGTGAAGCTATATTGGCCAGAGCAAGTGCTTCCGAAATAATGTTTCGTGAGGGATTTGACATTTCATTGCCATTATTTCATAAAGAACATCCAGAGAAAGGAGGAGTTCCACCTGCTGCTACTGTCAACCCATTTCCGGCTCAACTAACCACTTGCCGTCATGGAAAATCTTGGAAAGATTTAAGAGATGAAAGATGTGAAGAGGACAATAAAGAATATGACAAGTTTGATTATGAGCAGTTGTTAGCTAACTCAACTTTTTGCATTGTTGCACGAGGAAGACGATTAGGATCCTACAGATTTCTTGAAGCCTTAGCAGCTGGATGTATACCAGTTTTGCTAAGTAATGGATGGAGATTACCTTTTGATGAAAAAATAGATTGGAGGCGTGCAGTTATTTGGGCAGACGAACGCTTATTATTACAGGTTCCAGAGCTTGTACGTTCAGTACCACCGGAACGCATCCTTGCACTGAGACAGCAAACTCAGTTTTTATGGGAGCAATATTTTTCATCAATTGAAAAAATCGTCTTCACAACTATTGAGATTTTACTGGAACGTGTTCTCGCTCATCGAGTTTCAAAGCAACGAGATCTTCTAATTTGGAATGCTTCACCAGGAGCCCTCGGAACACTGGCTGCATATGGCGATTCTAGATCACATTACCCTATTTCATCTCCTGTATTAGAAGCGCCCTCGGCTCCTTTTACCAGCCTCGCCCCTCTTCCTGTTCCATCAATACCTCAGATGGCACCTCCTCACACTCCTGGTAATTCTTTTACTGCACTGTTATATGTTCAAGCAACATCACCAGCTTTACACAAACTCCTTGCTAATATTGCCAGTAGCGAGTTTTGCGAGAAAGTAGTTCTAGTATGGGACAGTGAACGGGCTGCGCCTTCTCTGAAGTCTTTGTCACGTATGGCTGGAGACAGCCGGGATCCACTCCCAGTAATAGTTGTCGACGCTACCACTCACTACCCTGGTGAAGGTGTTTCTGCACGATGGCAGCCATTGTGGGCTGTTCCGACCGCAGCTGTGTTTTCATTAGACGGCGACGCTCCACTTCTGGCTGAAGAATTGGATTTTGCTTTTCGCGTATGGCAACAATTCCCCGAACGTATTGTTGGGTATCCAGCTAGAACTCACTTTTGGGACGAATCAAAGGGGTCATGGGGTTACAGCAGTCGGTGGGGCAGCCTGTACTCCATGGTGCTGCCGGGTGCGGCACTAGTGCACCGCGCCACGCTGGCGTTGTACGCGGCGAGCGGGCCGGCGCTGCGGCAGGTCGTGCAGCGCGCGCACAACTGTGAGGACATCCTGCTCAACTGTCTCGTGGCGCACACCACGCGCCGCCCGCCGCTCAAGCTGGCTCAGCGCCGCCGCTACAAGCCGCCCCTCCATCACAAGTCGCTGTGGAGTGATCCTGAACATTTTGTTCAACGACAATCATGCTTGAACACATTTGCTACAACTTGGGGTTATATGCCACTGCTTCGATCGGTACTACGTCTCGACCCCATACTTTTCAAAGATCCCGTGTCTACCCTCAGGAAGAAATATCGAAAAATGGAATTAGTTTCATCTTAG
Protein
MDTCFDFSKCGSDPKIFVYPSDGTVSASYRKVLSVIRESRYVTRDPNEACLFVPAIDTLDADPLSSEHVSDVASRLSRLPYWRNGEAILARASASEIMFREGFDISLPLFHKEHPEKGGVPPAATVNPFPAQLTTCRHGKSWKDLRDERCEEDNKEYDKFDYEQLLANSTFCIVARGRRLGSYRFLEALAAGCIPVLLSNGWRLPFDEKIDWRRAVIWADERLLLQVPELVRSVPPERILALRQQTQFLWEQYFSSIEKIVFTTIEILLERVLAHRVSKQRDLLIWNASPGALGTLAAYGDSRSHYPISSPVLEAPSAPFTSLAPLPVPSIPQMAPPHTPGNSFTALLYVQATSPALHKLLANIASSEFCEKVVLVWDSERAAPSLKSLSRMAGDSRDPLPVIVVDATTHYPGEGVSARWQPLWAVPTAAVFSLDGDAPLLAEELDFAFRVWQQFPERIVGYPARTHFWDESKGSWGYSSRWGSLYSMVLPGAALVHRATLALYAASGPALRQVVQRAHNCEDILLNCLVAHTTRRPPLKLAQRRRYKPPLHHKSLWSDPEHFVQRQSCLNTFATTWGYMPLLRSVLRLDPILFKDPVSTLRKKYRKMELVSS
Summary
Description
Glycosyltransferase required for the biosynthesis of heparan-sulfate and responsible for the alternating addition of beta-1-4-linked glucuronic acid (GlcA) and alpha-1-4-linked N-acetylglucosamine (GlcNAc) units to nascent heparan sulfate chains. Botv is the trigger of heparan sulfate chain initiation and polymerization takes place by a complex of ttv and sotv. Plays a central role in the diffusion of morphogens hedgehog (hh), wingless (wg) and decapentaplegic (dpp) via its role in heparan sulfate proteoglycans (HSPGs) biosynthesis which are required for movement of hh, dpp and wg morphogens.
Catalytic Activity
3-O-{[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + UDP-N-acetyl-alpha-D-glucosamine = 3-O-{alpha-D-GlcNAc-[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + H(+) + UDP
3-O-{alpha-D-GlcNAc-[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-{[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n+1)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + H(+) + UDP
3-O-{alpha-D-GlcNAc-[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-{[(1->4)-beta-D-GlcA-(1->4)-alpha-D-GlcNAc](n+1)-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl}-L-seryl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Subunit
Interacts with sau (PubMed:23720043).
Miscellaneous
'Tout velu' means 'very hairy' in French.
Similarity
Belongs to the glycosyltransferase 47 family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Golgi apparatus
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Wnt signaling pathway
Feature
chain Exostosin-1
Uniprot
H9J2J0
A0A2A4JMK4
A0A2H1WVX4
A0A194RFY3
A0A194PQ24
A0A212FIZ7
+ More
A0A1B6I3V0 A0A1Y1KAS6 B3NR29 B4QFV3 A0A0B4KER9 B4P771 Q9V730 B4HRE3 A0A1W4X4F9 A0A182M694 N6TGI1 B3MII0 A0A0A9WWC0 A0A182R8X9 A0A1W4UJ84 A0A182YE64 A0A1L8E1L1 A0A182W8B0 A0A088AHS9 A0A2M4AMV9 A0A1S4H3W8 A0A0L7RIY1 A0A2M4CRW6 B4J686 A0A2R7W881 A0A182QQB2 A0A026WZB1 B4LLN6 A0A0N0BIM4 A0A069DW15 D6W7Y2 Q291D8 A0A3B0J1W9 A0A154P490 A0A023EZV3 E2BZD3 K7J1Q7 E2AJC3 A0A0J7P3A6 A0A1Q3FUS9 A0A195F1G4 B4KP04 E9IMK5 A0A151XCD1 A0A195E7J8 A0A195CH61 F4X6R0 A0A158NFT9 A0A0P5LKD7 A0A195B1Y1 A0A1S3CU44 A0A0P5TK58 A0A0P5QBK7 A0A0P5N8T4 A0A0P5NXH4 A0A0N8E558 A0A0P4ZZR8 A0A0P5P5G2 A0A0P5PSC2 A0A0P5RX86 A0A0P4Y8M4 A0A0N7ZX58 A0A0N8E6X1 A0A1J1ID21 A0A0P6EZE3 A0A232FJ93 A0A0P6ESH1 A0A0N8EEQ7 A0A0P5GSU3 E9GMR7 A0A0N8EDT1 E0VUU1 Q7QDS1 A0A2S2QCG4 A0A0P5ADS2 T1JM53 A0A0P5MDJ3 A0A1S3JTH4 A0A1Z5KUN5 A0A2R5LLN6 A0A131YVM5 A0A224YYF9 L7MJY8 A0A131XHI6 A0A3B3ZIL0 A0A131Y1V5 A0A2U9CWR0 W5UJD3 A0A147BVE1 A0A293M5E5 A0A3B4EFU2 A0A3B3BRZ6 W5KSV0 A0A1E1X3R6
A0A1B6I3V0 A0A1Y1KAS6 B3NR29 B4QFV3 A0A0B4KER9 B4P771 Q9V730 B4HRE3 A0A1W4X4F9 A0A182M694 N6TGI1 B3MII0 A0A0A9WWC0 A0A182R8X9 A0A1W4UJ84 A0A182YE64 A0A1L8E1L1 A0A182W8B0 A0A088AHS9 A0A2M4AMV9 A0A1S4H3W8 A0A0L7RIY1 A0A2M4CRW6 B4J686 A0A2R7W881 A0A182QQB2 A0A026WZB1 B4LLN6 A0A0N0BIM4 A0A069DW15 D6W7Y2 Q291D8 A0A3B0J1W9 A0A154P490 A0A023EZV3 E2BZD3 K7J1Q7 E2AJC3 A0A0J7P3A6 A0A1Q3FUS9 A0A195F1G4 B4KP04 E9IMK5 A0A151XCD1 A0A195E7J8 A0A195CH61 F4X6R0 A0A158NFT9 A0A0P5LKD7 A0A195B1Y1 A0A1S3CU44 A0A0P5TK58 A0A0P5QBK7 A0A0P5N8T4 A0A0P5NXH4 A0A0N8E558 A0A0P4ZZR8 A0A0P5P5G2 A0A0P5PSC2 A0A0P5RX86 A0A0P4Y8M4 A0A0N7ZX58 A0A0N8E6X1 A0A1J1ID21 A0A0P6EZE3 A0A232FJ93 A0A0P6ESH1 A0A0N8EEQ7 A0A0P5GSU3 E9GMR7 A0A0N8EDT1 E0VUU1 Q7QDS1 A0A2S2QCG4 A0A0P5ADS2 T1JM53 A0A0P5MDJ3 A0A1S3JTH4 A0A1Z5KUN5 A0A2R5LLN6 A0A131YVM5 A0A224YYF9 L7MJY8 A0A131XHI6 A0A3B3ZIL0 A0A131Y1V5 A0A2U9CWR0 W5UJD3 A0A147BVE1 A0A293M5E5 A0A3B4EFU2 A0A3B3BRZ6 W5KSV0 A0A1E1X3R6
Pubmed
19121390
26354079
22118469
28004739
17994087
22936249
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 9665133 16303756 10549295 10644674 10806213 12586063 14645127 14729575 14998928 15056609 23720043 23537049 25401762 26823975 25244985 12364791 24508170 26334808 18362917 19820115 15632085 25474469 20798317 20075255 21282665 21719571 21347285 28648823 21292972 20566863 28528879 26830274 28797301 25576852 28049606 25463417 23127152 29652888 29451363 25329095 28503490
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 9665133 16303756 10549295 10644674 10806213 12586063 14645127 14729575 14998928 15056609 23720043 23537049 25401762 26823975 25244985 12364791 24508170 26334808 18362917 19820115 15632085 25474469 20798317 20075255 21282665 21719571 21347285 28648823 21292972 20566863 28528879 26830274 28797301 25576852 28049606 25463417 23127152 29652888 29451363 25329095 28503490
EMBL
BABH01008008
BABH01008009
BABH01008010
BABH01008011
BABH01008012
NWSH01000990
+ More
PCG73211.1 ODYU01011465 SOQ57213.1 KQ460207 KPJ16723.1 KQ459597 KPI94839.1 AGBW02008321 OWR53704.1 GECU01026141 GECU01014269 GECU01000285 JAS81565.1 JAS93437.1 JAT07422.1 GEZM01087596 JAV58593.1 CH954179 EDV56018.1 CM000362 CM002911 EDX07098.1 KMY93784.1 AE013599 AGB93503.1 CM000158 EDW91036.1 AF083889 AB221351 BT021403 CH480816 EDW47872.1 AXCM01000199 APGK01039173 KB740967 KB632399 ENN76888.1 ERL94623.1 CH902619 EDV37028.1 GBHO01031565 GBRD01016843 GDHC01008020 JAG12039.1 JAG48984.1 JAQ10609.1 GFDF01001548 JAV12536.1 GGFK01008789 MBW42110.1 AAAB01008849 KQ414583 KOC70701.1 GGFL01003875 MBW68053.1 CH916367 EDW00859.1 KK854385 PTY15341.1 AXCN02002291 KK107063 EZA61138.1 CH940648 EDW60899.1 KQ435727 KOX77892.1 GBGD01000621 JAC88268.1 KQ971307 EFA11154.1 CM000071 EAL25174.2 OUUW01000001 SPP75304.1 KQ434806 KZC06144.1 GBBI01004391 JAC14321.1 GL451589 EFN78943.1 GL439972 EFN66444.1 LBMM01000226 KMR03929.1 GFDL01003833 JAV31212.1 KQ981864 KYN34308.1 CH933808 EDW10070.1 GL764195 EFZ18199.1 KQ982314 KYQ57970.1 KQ979568 KYN20827.1 KQ977754 KYN00065.1 GL888818 EGI57842.1 ADTU01014739 GDIQ01168360 JAK83365.1 KQ976662 KYM78486.1 GDIP01130734 JAL72980.1 GDIQ01265478 GDIQ01230080 GDIQ01227186 GDIQ01189157 GDIQ01173253 GDIQ01148475 GDIQ01135075 GDIQ01132873 GDIQ01130424 GDIQ01116380 GDIQ01109220 GDIQ01058548 GDIQ01049808 GDIQ01046703 GDIQ01032013 JAL35346.1 GDIQ01145622 JAL06104.1 GDIQ01254025 GDIQ01224837 GDIQ01207649 GDIQ01204185 GDIQ01202356 GDIQ01182958 GDIQ01181656 GDIQ01159671 GDIQ01157813 GDIQ01128834 GDIQ01100235 GDIQ01067298 GDIQ01041312 JAK93912.1 GDIQ01060804 JAN33933.1 GDIP01205970 JAJ17432.1 GDIQ01135119 JAL16607.1 GDIQ01146982 GDIQ01131624 GDIQ01130425 JAL21301.1 GDIQ01100236 JAL51490.1 GDIP01233271 JAI90130.1 GDIP01203873 JAJ19529.1 GDIQ01055900 JAN38837.1 CVRI01000047 CRK98107.1 GDIQ01055899 JAN38838.1 NNAY01000116 OXU30816.1 GDIQ01058547 JAN36190.1 GDIQ01034012 JAN60725.1 GDIQ01237332 JAK14393.1 GL732553 EFX79142.1 GDIQ01036620 JAN58117.1 DS235793 EEB17147.1 EAA07205.3 GGMS01006200 MBY75403.1 GDIP01213335 JAJ10067.1 JH432085 GDIQ01165424 JAK86301.1 GFJQ02008376 JAV98593.1 GGLE01006111 MBY10237.1 GEDV01005879 JAP82678.1 GFPF01007716 MAA18862.1 GACK01001456 JAA63578.1 GEFH01002018 JAP66563.1 GEFM01002567 JAP73229.1 CP026262 AWP20339.1 JT416729 AHH42126.1 GEGO01000681 JAR94723.1 GFWV01011464 MAA36193.1 GFAC01005296 JAT93892.1
PCG73211.1 ODYU01011465 SOQ57213.1 KQ460207 KPJ16723.1 KQ459597 KPI94839.1 AGBW02008321 OWR53704.1 GECU01026141 GECU01014269 GECU01000285 JAS81565.1 JAS93437.1 JAT07422.1 GEZM01087596 JAV58593.1 CH954179 EDV56018.1 CM000362 CM002911 EDX07098.1 KMY93784.1 AE013599 AGB93503.1 CM000158 EDW91036.1 AF083889 AB221351 BT021403 CH480816 EDW47872.1 AXCM01000199 APGK01039173 KB740967 KB632399 ENN76888.1 ERL94623.1 CH902619 EDV37028.1 GBHO01031565 GBRD01016843 GDHC01008020 JAG12039.1 JAG48984.1 JAQ10609.1 GFDF01001548 JAV12536.1 GGFK01008789 MBW42110.1 AAAB01008849 KQ414583 KOC70701.1 GGFL01003875 MBW68053.1 CH916367 EDW00859.1 KK854385 PTY15341.1 AXCN02002291 KK107063 EZA61138.1 CH940648 EDW60899.1 KQ435727 KOX77892.1 GBGD01000621 JAC88268.1 KQ971307 EFA11154.1 CM000071 EAL25174.2 OUUW01000001 SPP75304.1 KQ434806 KZC06144.1 GBBI01004391 JAC14321.1 GL451589 EFN78943.1 GL439972 EFN66444.1 LBMM01000226 KMR03929.1 GFDL01003833 JAV31212.1 KQ981864 KYN34308.1 CH933808 EDW10070.1 GL764195 EFZ18199.1 KQ982314 KYQ57970.1 KQ979568 KYN20827.1 KQ977754 KYN00065.1 GL888818 EGI57842.1 ADTU01014739 GDIQ01168360 JAK83365.1 KQ976662 KYM78486.1 GDIP01130734 JAL72980.1 GDIQ01265478 GDIQ01230080 GDIQ01227186 GDIQ01189157 GDIQ01173253 GDIQ01148475 GDIQ01135075 GDIQ01132873 GDIQ01130424 GDIQ01116380 GDIQ01109220 GDIQ01058548 GDIQ01049808 GDIQ01046703 GDIQ01032013 JAL35346.1 GDIQ01145622 JAL06104.1 GDIQ01254025 GDIQ01224837 GDIQ01207649 GDIQ01204185 GDIQ01202356 GDIQ01182958 GDIQ01181656 GDIQ01159671 GDIQ01157813 GDIQ01128834 GDIQ01100235 GDIQ01067298 GDIQ01041312 JAK93912.1 GDIQ01060804 JAN33933.1 GDIP01205970 JAJ17432.1 GDIQ01135119 JAL16607.1 GDIQ01146982 GDIQ01131624 GDIQ01130425 JAL21301.1 GDIQ01100236 JAL51490.1 GDIP01233271 JAI90130.1 GDIP01203873 JAJ19529.1 GDIQ01055900 JAN38837.1 CVRI01000047 CRK98107.1 GDIQ01055899 JAN38838.1 NNAY01000116 OXU30816.1 GDIQ01058547 JAN36190.1 GDIQ01034012 JAN60725.1 GDIQ01237332 JAK14393.1 GL732553 EFX79142.1 GDIQ01036620 JAN58117.1 DS235793 EEB17147.1 EAA07205.3 GGMS01006200 MBY75403.1 GDIP01213335 JAJ10067.1 JH432085 GDIQ01165424 JAK86301.1 GFJQ02008376 JAV98593.1 GGLE01006111 MBY10237.1 GEDV01005879 JAP82678.1 GFPF01007716 MAA18862.1 GACK01001456 JAA63578.1 GEFH01002018 JAP66563.1 GEFM01002567 JAP73229.1 CP026262 AWP20339.1 JT416729 AHH42126.1 GEGO01000681 JAR94723.1 GFWV01011464 MAA36193.1 GFAC01005296 JAT93892.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000008711
+ More
UP000000304 UP000000803 UP000002282 UP000001292 UP000192223 UP000075883 UP000019118 UP000030742 UP000007801 UP000075900 UP000192221 UP000076408 UP000075920 UP000005203 UP000053825 UP000001070 UP000075886 UP000053097 UP000008792 UP000053105 UP000007266 UP000001819 UP000268350 UP000076502 UP000008237 UP000002358 UP000000311 UP000036403 UP000078541 UP000009192 UP000075809 UP000078492 UP000078542 UP000007755 UP000005205 UP000078540 UP000079169 UP000183832 UP000215335 UP000000305 UP000009046 UP000007062 UP000085678 UP000261520 UP000246464 UP000221080 UP000261440 UP000261560 UP000018467
UP000000304 UP000000803 UP000002282 UP000001292 UP000192223 UP000075883 UP000019118 UP000030742 UP000007801 UP000075900 UP000192221 UP000076408 UP000075920 UP000005203 UP000053825 UP000001070 UP000075886 UP000053097 UP000008792 UP000053105 UP000007266 UP000001819 UP000268350 UP000076502 UP000008237 UP000002358 UP000000311 UP000036403 UP000078541 UP000009192 UP000075809 UP000078492 UP000078542 UP000007755 UP000005205 UP000078540 UP000079169 UP000183832 UP000215335 UP000000305 UP000009046 UP000007062 UP000085678 UP000261520 UP000246464 UP000221080 UP000261440 UP000261560 UP000018467
Interpro
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
H9J2J0
A0A2A4JMK4
A0A2H1WVX4
A0A194RFY3
A0A194PQ24
A0A212FIZ7
+ More
A0A1B6I3V0 A0A1Y1KAS6 B3NR29 B4QFV3 A0A0B4KER9 B4P771 Q9V730 B4HRE3 A0A1W4X4F9 A0A182M694 N6TGI1 B3MII0 A0A0A9WWC0 A0A182R8X9 A0A1W4UJ84 A0A182YE64 A0A1L8E1L1 A0A182W8B0 A0A088AHS9 A0A2M4AMV9 A0A1S4H3W8 A0A0L7RIY1 A0A2M4CRW6 B4J686 A0A2R7W881 A0A182QQB2 A0A026WZB1 B4LLN6 A0A0N0BIM4 A0A069DW15 D6W7Y2 Q291D8 A0A3B0J1W9 A0A154P490 A0A023EZV3 E2BZD3 K7J1Q7 E2AJC3 A0A0J7P3A6 A0A1Q3FUS9 A0A195F1G4 B4KP04 E9IMK5 A0A151XCD1 A0A195E7J8 A0A195CH61 F4X6R0 A0A158NFT9 A0A0P5LKD7 A0A195B1Y1 A0A1S3CU44 A0A0P5TK58 A0A0P5QBK7 A0A0P5N8T4 A0A0P5NXH4 A0A0N8E558 A0A0P4ZZR8 A0A0P5P5G2 A0A0P5PSC2 A0A0P5RX86 A0A0P4Y8M4 A0A0N7ZX58 A0A0N8E6X1 A0A1J1ID21 A0A0P6EZE3 A0A232FJ93 A0A0P6ESH1 A0A0N8EEQ7 A0A0P5GSU3 E9GMR7 A0A0N8EDT1 E0VUU1 Q7QDS1 A0A2S2QCG4 A0A0P5ADS2 T1JM53 A0A0P5MDJ3 A0A1S3JTH4 A0A1Z5KUN5 A0A2R5LLN6 A0A131YVM5 A0A224YYF9 L7MJY8 A0A131XHI6 A0A3B3ZIL0 A0A131Y1V5 A0A2U9CWR0 W5UJD3 A0A147BVE1 A0A293M5E5 A0A3B4EFU2 A0A3B3BRZ6 W5KSV0 A0A1E1X3R6
A0A1B6I3V0 A0A1Y1KAS6 B3NR29 B4QFV3 A0A0B4KER9 B4P771 Q9V730 B4HRE3 A0A1W4X4F9 A0A182M694 N6TGI1 B3MII0 A0A0A9WWC0 A0A182R8X9 A0A1W4UJ84 A0A182YE64 A0A1L8E1L1 A0A182W8B0 A0A088AHS9 A0A2M4AMV9 A0A1S4H3W8 A0A0L7RIY1 A0A2M4CRW6 B4J686 A0A2R7W881 A0A182QQB2 A0A026WZB1 B4LLN6 A0A0N0BIM4 A0A069DW15 D6W7Y2 Q291D8 A0A3B0J1W9 A0A154P490 A0A023EZV3 E2BZD3 K7J1Q7 E2AJC3 A0A0J7P3A6 A0A1Q3FUS9 A0A195F1G4 B4KP04 E9IMK5 A0A151XCD1 A0A195E7J8 A0A195CH61 F4X6R0 A0A158NFT9 A0A0P5LKD7 A0A195B1Y1 A0A1S3CU44 A0A0P5TK58 A0A0P5QBK7 A0A0P5N8T4 A0A0P5NXH4 A0A0N8E558 A0A0P4ZZR8 A0A0P5P5G2 A0A0P5PSC2 A0A0P5RX86 A0A0P4Y8M4 A0A0N7ZX58 A0A0N8E6X1 A0A1J1ID21 A0A0P6EZE3 A0A232FJ93 A0A0P6ESH1 A0A0N8EEQ7 A0A0P5GSU3 E9GMR7 A0A0N8EDT1 E0VUU1 Q7QDS1 A0A2S2QCG4 A0A0P5ADS2 T1JM53 A0A0P5MDJ3 A0A1S3JTH4 A0A1Z5KUN5 A0A2R5LLN6 A0A131YVM5 A0A224YYF9 L7MJY8 A0A131XHI6 A0A3B3ZIL0 A0A131Y1V5 A0A2U9CWR0 W5UJD3 A0A147BVE1 A0A293M5E5 A0A3B4EFU2 A0A3B3BRZ6 W5KSV0 A0A1E1X3R6
PDB
1ON8
E-value=1.27519e-22,
Score=265
Ontologies
PATHWAY
GO
GO:0006486
GO:0016757
GO:0016021
GO:0008354
GO:0090098
GO:0050509
GO:0045880
GO:0005794
GO:0090263
GO:0015014
GO:0008375
GO:0005783
GO:0007411
GO:0030210
GO:0050508
GO:0008589
GO:0016055
GO:0000139
GO:0006044
GO:0005789
GO:0015012
GO:0046872
GO:0006024
GO:0015020
GO:0006418
GO:0004823
GO:0006429
GO:0006631
GO:0006635
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
Golgi apparatus membrane
Golgi apparatus membrane
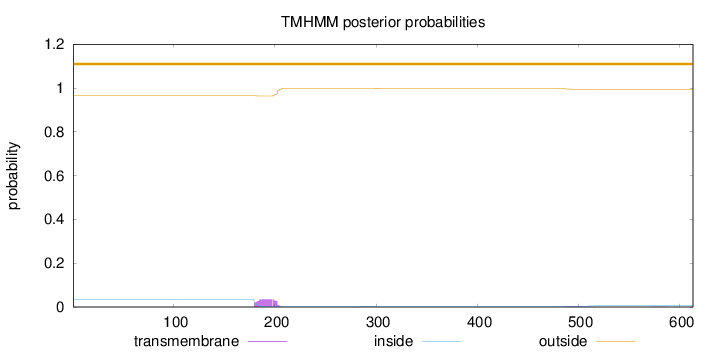
Length:
613
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.866819999999998
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.03561
outside
1 - 613
Population Genetic Test Statistics
Pi
256.296978
Theta
199.319842
Tajima's D
0.894397
CLR
0.220094
CSRT
0.632168391580421
Interpretation
Uncertain
