Pre Gene Modal
BGIBMGA003466
Annotation
PREDICTED:_FUN14_domain-containing_protein_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.962
Sequence
CDS
ATGGCCAAACCTAAGAACGAAGATGCTTCTGAGGAAGCTAAAAAAATTGTAGACGACGCTAAAAACTTTATCGAGAGAGCCATTGCAGATATTGGTAAAACTTCAGCAACTAAGCAGCTAATTTTAGGAACTGCATCAGGATGGATAACTGGTTTCATTTCTATGAAGATTGGAAAAGTAGCAGCTGTTGGACTTGGAGGTGGAGTCATTCTTCTACACATTGCAAGCCAAAAAGGCTACATTGATATTAATTGGGATAAAATAAACAAAAAAGTTGATAAGATCAGTGATAAAATTGAAAAAGAAGCTACTGGAAAATCCCCAGATTGGTTTGAAAAGGTGGAAAGATTTGTAGACCGAAAATTGGATAAAGCTGAAGAACTTATAAAAAAGAAGGAAATAAAGGCAAAAAAATGGTATAATAATTTCACTGGTGATGACCAATACCGTGCTACAGAATCACATGTCTTTTTGGCAGCATTTTTTGCTGGAATGGCTATTGGTCTAATTTGCGGAAAGTAA
Protein
MAKPKNEDASEEAKKIVDDAKNFIERAIADIGKTSATKQLILGTASGWITGFISMKIGKVAAVGLGGGVILLHIASQKGYIDINWDKINKKVDKISDKIEKEATGKSPDWFEKVERFVDRKLDKAEELIKKKEIKAKKWYNNFTGDDQYRATESHVFLAAFFAGMAIGLICGK
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
H9J1S9
A0A194RL62
A0A194PPD2
A0A2A4JI03
S4PGY3
A0A0L7KRJ6
+ More
A0A2H1WQV5 A0A1E1VYR0 A0A212FIZ4 A0A1W4WZP1 A0A2P8Z3E8 A0A1Y1MQ03 A0A158NMG3 A0A195CVQ9 A0A232FF40 E1ZZ50 F4WI55 A0A087ZPB7 A0A2A3E637 E2BQG3 V5GFN8 T1DTR7 A0A182Q9A4 A0A1S4G391 B0W009 T1DQ09 A0A1Q3FKT2 A0A2J7QAL0 A0A182Y8F6 Q16IJ5 A0A182PAT2 A0A1S4GYG0 A0A2C9GS54 A0A182UMY1 A0A182TXF0 A0A2M4ABG6 A0A182K038 W5JC15 A0A2M3Z4X0 A0A2M4C254 Q16FE9 A0A182J4H3 A0A2M4C184 A0A0N0BIT6 A0A1Q3FKJ8 A0A1Q3FKM3 A0A1B6FGB5 A0A154NZY9 A0A023EK56 A0A182MGI8 A0A182VTR6 A0A1I8JUL9 A0A1L8DUU5 A0A1L8DW64 A0A1B6HAU8 A0A1B6KJ32 A0A084VNK9 A0A026WFA2 A0A067RBX6 A0A0T6B4V8 A0A1B0FY91 A0A1J1INP7 C4WSD0 A0A182S638 A0A3L8DKR2 A0A0L7R419 A0A2S2RA86 E9IEH5 A0A182NB25 A0A0L0BZB8 A0A2H8TEJ5 A0A151JVF4 A0A0K8SUU4 A0A1B0GGW4 W8AHQ2 A0A336MKQ7 A0A336MZU3 A0A034WMA4 A0A0A1X1Y0 A0A1A9ZVR2 D3TLS4 A0A1A9XJC5 A0A1B0BLP0 A0A1A9URM5 B3N950 A0A1Y1MJZ8 E0W4E6 A0A182X167 A0A1I8P8P5 A0A1L8EEE9 B4KJH5 B4JPL8 A0A1W4UT19 A0A1I8MMN1 B4NZ74
A0A2H1WQV5 A0A1E1VYR0 A0A212FIZ4 A0A1W4WZP1 A0A2P8Z3E8 A0A1Y1MQ03 A0A158NMG3 A0A195CVQ9 A0A232FF40 E1ZZ50 F4WI55 A0A087ZPB7 A0A2A3E637 E2BQG3 V5GFN8 T1DTR7 A0A182Q9A4 A0A1S4G391 B0W009 T1DQ09 A0A1Q3FKT2 A0A2J7QAL0 A0A182Y8F6 Q16IJ5 A0A182PAT2 A0A1S4GYG0 A0A2C9GS54 A0A182UMY1 A0A182TXF0 A0A2M4ABG6 A0A182K038 W5JC15 A0A2M3Z4X0 A0A2M4C254 Q16FE9 A0A182J4H3 A0A2M4C184 A0A0N0BIT6 A0A1Q3FKJ8 A0A1Q3FKM3 A0A1B6FGB5 A0A154NZY9 A0A023EK56 A0A182MGI8 A0A182VTR6 A0A1I8JUL9 A0A1L8DUU5 A0A1L8DW64 A0A1B6HAU8 A0A1B6KJ32 A0A084VNK9 A0A026WFA2 A0A067RBX6 A0A0T6B4V8 A0A1B0FY91 A0A1J1INP7 C4WSD0 A0A182S638 A0A3L8DKR2 A0A0L7R419 A0A2S2RA86 E9IEH5 A0A182NB25 A0A0L0BZB8 A0A2H8TEJ5 A0A151JVF4 A0A0K8SUU4 A0A1B0GGW4 W8AHQ2 A0A336MKQ7 A0A336MZU3 A0A034WMA4 A0A0A1X1Y0 A0A1A9ZVR2 D3TLS4 A0A1A9XJC5 A0A1B0BLP0 A0A1A9URM5 B3N950 A0A1Y1MJZ8 E0W4E6 A0A182X167 A0A1I8P8P5 A0A1L8EEE9 B4KJH5 B4JPL8 A0A1W4UT19 A0A1I8MMN1 B4NZ74
Pubmed
EMBL
BABH01008013
KQ460207
KPJ16721.1
KQ459597
KPI94838.1
NWSH01001393
+ More
PCG71406.1 GAIX01006010 JAA86550.1 JTDY01006845 KOB65651.1 ODYU01010383 SOQ55461.1 GDQN01011243 GDQN01005301 GDQN01003419 GDQN01001107 JAT79811.1 JAT85753.1 JAT87635.1 JAT89947.1 AGBW02008321 OWR53706.1 PYGN01000215 PSN51026.1 GEZM01029623 JAV86036.1 ADTU01020462 KQ977231 KYN04761.1 NNAY01000298 OXU29374.1 GL435293 EFN73541.1 GL888172 EGI66148.1 KZ288381 PBC26521.1 GL449769 EFN82021.1 GALX01005667 JAB62799.1 GAMD01000789 JAB00802.1 AXCN02000647 DS231816 EDS37581.1 GAMD01002859 JAA98731.1 GFDL01006877 JAV28168.1 NEVH01016330 PNF25612.1 CH478073 EAT34088.1 AAAB01008964 APCN01000749 GGFK01004813 MBW38134.1 ADMH02001646 ETN61611.1 GGFM01002820 MBW23571.1 GGFJ01009927 MBW59068.1 CH478458 EAT32960.1 GGFJ01009926 MBW59067.1 KQ435726 KOX78142.1 GFDL01006938 JAV28107.1 GFDL01006937 JAV28108.1 GECZ01020595 JAS49174.1 KQ434786 KZC05152.1 GAPW01004096 JAC09502.1 AXCM01000962 GFDF01003883 JAV10201.1 GFDF01003412 JAV10672.1 GECU01035934 JAS71772.1 GEBQ01028515 JAT11462.1 ATLV01014760 KE524984 KFB39553.1 KK107238 EZA54800.1 KK852804 KDR16193.1 LJIG01009793 KRT82370.1 AJVK01000819 CVRI01000056 CRL01784.1 ABLF02017409 AK340143 AK340144 BAH70800.1 QOIP01000007 RLU21020.1 KQ414663 KOC65496.1 GGMS01017377 MBY86580.1 GL762591 EFZ21049.1 JRES01001123 KNC25336.1 GFXV01000722 MBW12527.1 KQ981692 KYN37885.1 GBRD01009166 GDHC01008811 JAG56655.1 JAQ09818.1 AJWK01000780 AJWK01020170 GAMC01018555 JAB88000.1 UFQT01001425 SSX30500.1 UFQS01002870 UFQT01002870 SSX14802.1 SSX34193.1 GAKP01003662 JAC55290.1 GBXI01008978 JAD05314.1 CCAG010009466 EZ422376 ADD18652.1 JXJN01016521 CH954177 EDV58485.1 GEZM01029625 JAV86034.1 DS235886 EEB20502.1 GFDG01001712 JAV17087.1 CH933807 EDW13555.1 CH916372 EDV98848.1 CM000157 EDW88769.1
PCG71406.1 GAIX01006010 JAA86550.1 JTDY01006845 KOB65651.1 ODYU01010383 SOQ55461.1 GDQN01011243 GDQN01005301 GDQN01003419 GDQN01001107 JAT79811.1 JAT85753.1 JAT87635.1 JAT89947.1 AGBW02008321 OWR53706.1 PYGN01000215 PSN51026.1 GEZM01029623 JAV86036.1 ADTU01020462 KQ977231 KYN04761.1 NNAY01000298 OXU29374.1 GL435293 EFN73541.1 GL888172 EGI66148.1 KZ288381 PBC26521.1 GL449769 EFN82021.1 GALX01005667 JAB62799.1 GAMD01000789 JAB00802.1 AXCN02000647 DS231816 EDS37581.1 GAMD01002859 JAA98731.1 GFDL01006877 JAV28168.1 NEVH01016330 PNF25612.1 CH478073 EAT34088.1 AAAB01008964 APCN01000749 GGFK01004813 MBW38134.1 ADMH02001646 ETN61611.1 GGFM01002820 MBW23571.1 GGFJ01009927 MBW59068.1 CH478458 EAT32960.1 GGFJ01009926 MBW59067.1 KQ435726 KOX78142.1 GFDL01006938 JAV28107.1 GFDL01006937 JAV28108.1 GECZ01020595 JAS49174.1 KQ434786 KZC05152.1 GAPW01004096 JAC09502.1 AXCM01000962 GFDF01003883 JAV10201.1 GFDF01003412 JAV10672.1 GECU01035934 JAS71772.1 GEBQ01028515 JAT11462.1 ATLV01014760 KE524984 KFB39553.1 KK107238 EZA54800.1 KK852804 KDR16193.1 LJIG01009793 KRT82370.1 AJVK01000819 CVRI01000056 CRL01784.1 ABLF02017409 AK340143 AK340144 BAH70800.1 QOIP01000007 RLU21020.1 KQ414663 KOC65496.1 GGMS01017377 MBY86580.1 GL762591 EFZ21049.1 JRES01001123 KNC25336.1 GFXV01000722 MBW12527.1 KQ981692 KYN37885.1 GBRD01009166 GDHC01008811 JAG56655.1 JAQ09818.1 AJWK01000780 AJWK01020170 GAMC01018555 JAB88000.1 UFQT01001425 SSX30500.1 UFQS01002870 UFQT01002870 SSX14802.1 SSX34193.1 GAKP01003662 JAC55290.1 GBXI01008978 JAD05314.1 CCAG010009466 EZ422376 ADD18652.1 JXJN01016521 CH954177 EDV58485.1 GEZM01029625 JAV86034.1 DS235886 EEB20502.1 GFDG01001712 JAV17087.1 CH933807 EDW13555.1 CH916372 EDV98848.1 CM000157 EDW88769.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000192223 UP000245037 UP000005205 UP000078542 UP000215335 UP000000311 UP000007755 UP000005203 UP000242457 UP000008237 UP000075886 UP000002320 UP000235965 UP000076408 UP000008820 UP000075885 UP000075840 UP000075903 UP000075902 UP000075881 UP000000673 UP000075880 UP000053105 UP000076502 UP000075883 UP000075920 UP000075900 UP000030765 UP000053097 UP000027135 UP000092462 UP000183832 UP000007819 UP000075901 UP000279307 UP000053825 UP000075884 UP000037069 UP000078541 UP000092461 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000008711 UP000009046 UP000076407 UP000095300 UP000009192 UP000001070 UP000192221 UP000095301 UP000002282
UP000192223 UP000245037 UP000005205 UP000078542 UP000215335 UP000000311 UP000007755 UP000005203 UP000242457 UP000008237 UP000075886 UP000002320 UP000235965 UP000076408 UP000008820 UP000075885 UP000075840 UP000075903 UP000075902 UP000075881 UP000000673 UP000075880 UP000053105 UP000076502 UP000075883 UP000075920 UP000075900 UP000030765 UP000053097 UP000027135 UP000092462 UP000183832 UP000007819 UP000075901 UP000279307 UP000053825 UP000075884 UP000037069 UP000078541 UP000092461 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000008711 UP000009046 UP000076407 UP000095300 UP000009192 UP000001070 UP000192221 UP000095301 UP000002282
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J1S9
A0A194RL62
A0A194PPD2
A0A2A4JI03
S4PGY3
A0A0L7KRJ6
+ More
A0A2H1WQV5 A0A1E1VYR0 A0A212FIZ4 A0A1W4WZP1 A0A2P8Z3E8 A0A1Y1MQ03 A0A158NMG3 A0A195CVQ9 A0A232FF40 E1ZZ50 F4WI55 A0A087ZPB7 A0A2A3E637 E2BQG3 V5GFN8 T1DTR7 A0A182Q9A4 A0A1S4G391 B0W009 T1DQ09 A0A1Q3FKT2 A0A2J7QAL0 A0A182Y8F6 Q16IJ5 A0A182PAT2 A0A1S4GYG0 A0A2C9GS54 A0A182UMY1 A0A182TXF0 A0A2M4ABG6 A0A182K038 W5JC15 A0A2M3Z4X0 A0A2M4C254 Q16FE9 A0A182J4H3 A0A2M4C184 A0A0N0BIT6 A0A1Q3FKJ8 A0A1Q3FKM3 A0A1B6FGB5 A0A154NZY9 A0A023EK56 A0A182MGI8 A0A182VTR6 A0A1I8JUL9 A0A1L8DUU5 A0A1L8DW64 A0A1B6HAU8 A0A1B6KJ32 A0A084VNK9 A0A026WFA2 A0A067RBX6 A0A0T6B4V8 A0A1B0FY91 A0A1J1INP7 C4WSD0 A0A182S638 A0A3L8DKR2 A0A0L7R419 A0A2S2RA86 E9IEH5 A0A182NB25 A0A0L0BZB8 A0A2H8TEJ5 A0A151JVF4 A0A0K8SUU4 A0A1B0GGW4 W8AHQ2 A0A336MKQ7 A0A336MZU3 A0A034WMA4 A0A0A1X1Y0 A0A1A9ZVR2 D3TLS4 A0A1A9XJC5 A0A1B0BLP0 A0A1A9URM5 B3N950 A0A1Y1MJZ8 E0W4E6 A0A182X167 A0A1I8P8P5 A0A1L8EEE9 B4KJH5 B4JPL8 A0A1W4UT19 A0A1I8MMN1 B4NZ74
A0A2H1WQV5 A0A1E1VYR0 A0A212FIZ4 A0A1W4WZP1 A0A2P8Z3E8 A0A1Y1MQ03 A0A158NMG3 A0A195CVQ9 A0A232FF40 E1ZZ50 F4WI55 A0A087ZPB7 A0A2A3E637 E2BQG3 V5GFN8 T1DTR7 A0A182Q9A4 A0A1S4G391 B0W009 T1DQ09 A0A1Q3FKT2 A0A2J7QAL0 A0A182Y8F6 Q16IJ5 A0A182PAT2 A0A1S4GYG0 A0A2C9GS54 A0A182UMY1 A0A182TXF0 A0A2M4ABG6 A0A182K038 W5JC15 A0A2M3Z4X0 A0A2M4C254 Q16FE9 A0A182J4H3 A0A2M4C184 A0A0N0BIT6 A0A1Q3FKJ8 A0A1Q3FKM3 A0A1B6FGB5 A0A154NZY9 A0A023EK56 A0A182MGI8 A0A182VTR6 A0A1I8JUL9 A0A1L8DUU5 A0A1L8DW64 A0A1B6HAU8 A0A1B6KJ32 A0A084VNK9 A0A026WFA2 A0A067RBX6 A0A0T6B4V8 A0A1B0FY91 A0A1J1INP7 C4WSD0 A0A182S638 A0A3L8DKR2 A0A0L7R419 A0A2S2RA86 E9IEH5 A0A182NB25 A0A0L0BZB8 A0A2H8TEJ5 A0A151JVF4 A0A0K8SUU4 A0A1B0GGW4 W8AHQ2 A0A336MKQ7 A0A336MZU3 A0A034WMA4 A0A0A1X1Y0 A0A1A9ZVR2 D3TLS4 A0A1A9XJC5 A0A1B0BLP0 A0A1A9URM5 B3N950 A0A1Y1MJZ8 E0W4E6 A0A182X167 A0A1I8P8P5 A0A1L8EEE9 B4KJH5 B4JPL8 A0A1W4UT19 A0A1I8MMN1 B4NZ74
Ontologies
PANTHER
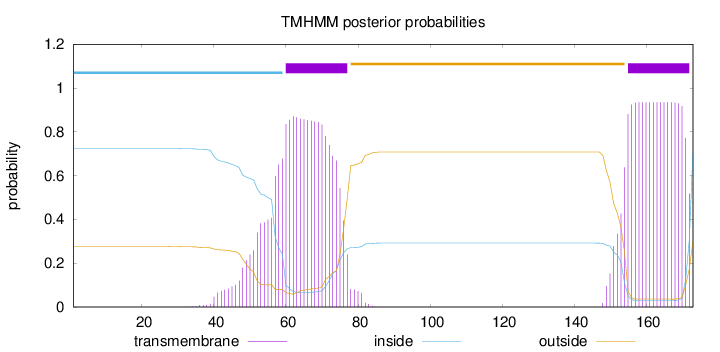
Topology
Length:
173
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
37.31177
Exp number, first 60 AAs:
6.28491
Total prob of N-in:
0.72389
inside
1 - 59
TMhelix
60 - 77
outside
78 - 154
TMhelix
155 - 172
inside
173 - 173
Population Genetic Test Statistics
Pi
137.67035
Theta
198.536662
Tajima's D
-1.305191
CLR
149.803623
CSRT
0.0894955252237388
Interpretation
Uncertain
