Pre Gene Modal
BGIBMGA003731
Annotation
PREDICTED:_potassium_voltage-gated_channel_subfamily_KQT_member_2_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.33
Sequence
CDS
ATGATTGTGTTTTTGATGGTGTTCATGTGTTTGTCGCTGTCTGTGTTCTCCACAATCGAAGAGTACGAAGAGAAAGCGGGGATCGTTCTCTTCTACATGGAGACAGTGGTGGTTGTATGGTTCGCTATTGAATTCATTTTAAGGCTATGGTCGTCAGGATGCAGGTCACGATATCAAGGATCGATAGGCCGCCTCAAATTTTTGCGGCGGCCTTTTTGCATTATAGACGTTACTACGATACTGGCGTCGTTGGTGGTTCTGGGTATGGGTTCGTCGAAGCAGGTTTTCGCGGCGTCAGCGTTGCGCGGCCTGCGATTCTTCCAGATACTTCGAATGGTTCGCATGGATCGTCGAGGTGGTACATGGAAACTACTAGGATCAGTCGTTTACGCGCATAGACAGGAATTAATAACGACTTTATACATAGGTTTCCTTGGATTGATATTTGCATCGTTTCTAGTCTATCTTGCTGAAAAAGATGTAGCCGATACCAAATTTAAAAACTTTGCTGAGGCTTTGTGGTGGGGAGTGATCACGCTGTGCACGGTGGGATATGGCGACATGGTGCCGCAGACCTGGCAGGGTAAATTCATTGCGTCTTTTTGTGCTCTGCTTGGGATATCATTCTTTGCCCTTCCTGCGGGTATTTTAGGTTCGGGTTTCGCTTTGAAAGTTCAGCAACAGCAACGTCAAAAGCACATGATAAGGAGAAGACAGCCAGCGGCAACACTCATACAGGCTTTGTGGAGATGCTACGCGGCTGATGAGCATTCCATGAGCGTTGCTACTTGGAAAATACACCAGATCCCGTTGCCTAGTCCTCAAACAAGTCGAGTGATGAGCGCGTCATTCAAGAACAATGCATCCTTTGTGTCGCGGTTGCCGACGATCCGACGGCACAAGACCACGTCCCTGCACTCCCCCGCGCCGCACTCCAAGCCGGCGCACAGATACGACGTTAACGCTAGCGCTGAAAATCTCGATGAGGAAGATGAGCCACGCTGTGTTACTCTAACGCCGCGACACAAAGCTGCAATACGTTTTATTAGAAAGCTCAAGTATTTTGTCGCACGCAAGAGATTTAGAGAAGCGTTGAAACCTTACGACGTGAAAGACGTGATAGAGCAATACTCTTCGGGACATGCGGATCTGCTAAACCGAACCAAGAATTTACAGTACAGGTTAGACCAAATACTTGGCAAACAAGGGTCAAAGGCTAAAGACGTCTACGCATCCAAAATAAGTCTTGCTTCCAGAGTAGTTAAAATCGAAAGACAAGTAGATGACATAGAGAGTAAGTTGGATCATTTATTAGAGATGTATGAAGAAGATAGAAGATCGCATAGGGCAGGTGGTGGAAGGAATGGAGGTGAGGGTGGGCCTTCAGGAGAATGTGGATTAGCGCTAAGAATGCGACCGGCTTTGTCAGACAAACAGTGCTCGGAGCCGAACTCTCCAGTATCTCGAACTTTCGACCAACCGGCTCCGGCATCGGCACCTCCTCTTCCTCATAAACGACCTATGAACAGAGGATATTCAGATTTGGGAACTAGGTTTATGAGGAAAAAAGTGATTGTTAAAACATCGTCTAGTCGGAACAGTGATTCCCCCAGAGACGATGAGGTAGTTATCGTAGTTCCCACAGACAGAGAGGACACGCCACCGCGACTTGTCACCGACAGCTCAGAAGTGTGCGCTAGCTGTTCGGTGGAAGTGGAGACAGAGGCTGATGTGGGTGGATCCCATTCTGGAAGTTCACCATCAGCATCTTGGTGCGAGGGTGATGGCACCGAAGAGCGCGGTTACGGCTCCGGGGACTCTCTGGCCGAGGATACCGCACTTCTGGGTGACGCGGCGCAGTCTGCCAACCCGCCGCGGATAGTCCGGAACCAGCGCCGGGACGTAGCCGTCGATCTCCACCTCCTACGACCTGATGTTTGA
Protein
MIVFLMVFMCLSLSVFSTIEEYEEKAGIVLFYMETVVVVWFAIEFILRLWSSGCRSRYQGSIGRLKFLRRPFCIIDVTTILASLVVLGMGSSKQVFAASALRGLRFFQILRMVRMDRRGGTWKLLGSVVYAHRQELITTLYIGFLGLIFASFLVYLAEKDVADTKFKNFAEALWWGVITLCTVGYGDMVPQTWQGKFIASFCALLGISFFALPAGILGSGFALKVQQQQRQKHMIRRRQPAATLIQALWRCYAADEHSMSVATWKIHQIPLPSPQTSRVMSASFKNNASFVSRLPTIRRHKTTSLHSPAPHSKPAHRYDVNASAENLDEEDEPRCVTLTPRHKAAIRFIRKLKYFVARKRFREALKPYDVKDVIEQYSSGHADLLNRTKNLQYRLDQILGKQGSKAKDVYASKISLASRVVKIERQVDDIESKLDHLLEMYEEDRRSHRAGGGRNGGEGGPSGECGLALRMRPALSDKQCSEPNSPVSRTFDQPAPASAPPLPHKRPMNRGYSDLGTRFMRKKVIVKTSSSRNSDSPRDDEVVIVVPTDREDTPPRLVTDSSEVCASCSVEVETEADVGGSHSGSSPSASWCEGDGTEERGYGSGDSLAEDTALLGDAAQSANPPRIVRNQRRDVAVDLHLLRPDV
Summary
Similarity
Belongs to the potassium channel family.
Uniprot
H9J2J4
A0A2A4JJ12
A0A2A4JHX1
A0A212FJ06
A0A1Q3FUE3
A0A1Q3FUT6
+ More
A0A1S4GRV7 D6W7C2 A0A1I8JU81 A7UU78 A0A084WEL6 A0A1S4GRV3 U5EU00 Q7PP97 A0A1Q3FUC3 A0A1L8DDI5 A0A182WQL0 A0A0C9Q7B4 A0A1Q3FTL0 A0A1Q3FUI8 A0A1Q3FUV2 K7IPU7 A0A1L8DDG6 W8B9C3 W8C0T6 A0A1I8NH71 A0A1Q3FUE7 A0A0Q9W4V5 A0A0P8XMR1 A0A0A1WIA3 E0VXL6 A0A1I8PAH7 A0A1L8DDH4 A0A0J9R9I6 A0A0B4KEI9 W8B271 A0A0R1DMY3 A0A0Q5WA09 A0A1W4VQP6 A0A0K8W891 A0A1W4VCN5 A0A0L7L344 T1PAX7 A0A0Q9W5H4 A0A0Q9WGP0 A0A0P8ZMI0 A0A0P8XMV7 A0A0N8P028 A0A0J9RAH7 A0A0P9AD25 A0A0Q9W508 A0A1I8NH63 A0A0R3NMI1 B7YZR3 A0A0R1DTU8 A0A0Q5WAC6 A0A0B4KF62 A0A0J9TXZ2 A0A3B0JLF9 Q5PXF9 A0A0R1DTZ7 A0A0Q5W9T9 A0A1W4VPD3 A0A0J9TY09 Q8IT87 H1UUH4 A0A0R1DU92 A0A0J9R9G6 A0A0B4KFJ4 A0A0Q9X0M2 A0A1W4VCN0 Q8IGU8 A0A0Q5W9Y9 A0A1W4VQQ1 B7YZR4 A0A0J9TXZ7 W8BE11 A0A0R1DPY4 A0A0Q5WM09 A0A1W4VQ35 A0A1Q3FTX1 A0A0Q9WGI4 A0A1I8NH75 A0A0R3NMX7 A0A0P9BWT0 A0A0R3NT21 A0A0R3NMK4 A0A0J9RAI8 A0A0B4KEP9 A0A0R1DTY9 A0A0J9R9G0 A0A0Q5WGU4 A0A0Q9WFP3 A0A1W4VQ39 A0A0P8YA19 A0A0J9TY03 A0A0B4KEH2 A0A0Q5WLK2 A0A0R1DMT9 A0A1W4VCC6
A0A1S4GRV7 D6W7C2 A0A1I8JU81 A7UU78 A0A084WEL6 A0A1S4GRV3 U5EU00 Q7PP97 A0A1Q3FUC3 A0A1L8DDI5 A0A182WQL0 A0A0C9Q7B4 A0A1Q3FTL0 A0A1Q3FUI8 A0A1Q3FUV2 K7IPU7 A0A1L8DDG6 W8B9C3 W8C0T6 A0A1I8NH71 A0A1Q3FUE7 A0A0Q9W4V5 A0A0P8XMR1 A0A0A1WIA3 E0VXL6 A0A1I8PAH7 A0A1L8DDH4 A0A0J9R9I6 A0A0B4KEI9 W8B271 A0A0R1DMY3 A0A0Q5WA09 A0A1W4VQP6 A0A0K8W891 A0A1W4VCN5 A0A0L7L344 T1PAX7 A0A0Q9W5H4 A0A0Q9WGP0 A0A0P8ZMI0 A0A0P8XMV7 A0A0N8P028 A0A0J9RAH7 A0A0P9AD25 A0A0Q9W508 A0A1I8NH63 A0A0R3NMI1 B7YZR3 A0A0R1DTU8 A0A0Q5WAC6 A0A0B4KF62 A0A0J9TXZ2 A0A3B0JLF9 Q5PXF9 A0A0R1DTZ7 A0A0Q5W9T9 A0A1W4VPD3 A0A0J9TY09 Q8IT87 H1UUH4 A0A0R1DU92 A0A0J9R9G6 A0A0B4KFJ4 A0A0Q9X0M2 A0A1W4VCN0 Q8IGU8 A0A0Q5W9Y9 A0A1W4VQQ1 B7YZR4 A0A0J9TXZ7 W8BE11 A0A0R1DPY4 A0A0Q5WM09 A0A1W4VQ35 A0A1Q3FTX1 A0A0Q9WGI4 A0A1I8NH75 A0A0R3NMX7 A0A0P9BWT0 A0A0R3NT21 A0A0R3NMK4 A0A0J9RAI8 A0A0B4KEP9 A0A0R1DTY9 A0A0J9R9G0 A0A0Q5WGU4 A0A0Q9WFP3 A0A1W4VQ39 A0A0P8YA19 A0A0J9TY03 A0A0B4KEH2 A0A0Q5WLK2 A0A0R1DMT9 A0A1W4VCC6
Pubmed
EMBL
BABH01008023
BABH01008024
BABH01008025
BABH01008026
NWSH01001393
PCG71400.1
+ More
PCG71399.1 AGBW02008321 OWR53709.1 GFDL01003825 JAV31220.1 GFDL01003823 JAV31222.1 AAAB01008960 KQ971307 EFA11047.2 EDO63856.1 ATLV01023225 ATLV01023226 ATLV01023227 KE525341 KFB48660.1 GANO01004123 JAB55748.1 EAA10809.4 GFDL01003824 JAV31221.1 GFDF01009572 JAV04512.1 GBYB01010048 JAG79815.1 GFDL01004111 JAV30934.1 GFDL01003820 JAV31225.1 GFDL01003822 JAV31223.1 GFDF01009571 JAV04513.1 GAMC01011318 JAB95237.1 GAMC01011321 JAB95234.1 GFDL01003821 JAV31224.1 CH940648 KRF80153.1 CH902619 KPU75962.1 GBXI01015533 GBXI01010827 JAC98758.1 JAD03465.1 DS235833 EEB18122.1 GFDF01009582 JAV04502.1 CM002911 KMY92707.1 AE013599 AGB93382.1 GAMC01011320 JAB95235.1 CM000157 KRJ98588.1 CH954177 KQS70284.1 GDHF01004891 JAI47423.1 JTDY01003220 KOB69928.1 KA645887 AFP60516.1 KRF80156.1 KRF80157.1 KRF80155.1 KPU75967.1 KPU75963.1 KPU75964.1 KMY92709.1 KPU75961.1 KRF80154.1 CM000071 KRT02122.1 ACL83086.1 KRJ98592.1 KQS70292.1 AGB93381.1 KMY92705.1 OUUW01000001 SPP74076.1 AY823300 AY823301 AAF58830.4 AAV70986.1 AAV70987.1 KRJ98593.1 KQS70289.1 KMY92720.1 AF536215 AAN63887.3 BT133059 AEX93144.1 KRJ98595.1 KMY92718.1 AGB93385.1 CH963719 KRF97437.1 BT001588 AAN71343.1 KQS70291.1 ACL83085.1 KMY92710.1 GAMC01011322 JAB95233.1 KRJ98589.1 KQS70282.1 GFDL01004119 JAV30926.1 KRF80158.1 KRT02124.1 KPU75966.1 KRT02133.1 KRT02121.1 KMY92719.1 AGB93384.1 KRJ98585.1 KMY92713.1 KQS70293.1 KRF80159.1 KPU75965.1 KMY92715.1 AGB93383.1 KQS70287.1 KRJ98586.1
PCG71399.1 AGBW02008321 OWR53709.1 GFDL01003825 JAV31220.1 GFDL01003823 JAV31222.1 AAAB01008960 KQ971307 EFA11047.2 EDO63856.1 ATLV01023225 ATLV01023226 ATLV01023227 KE525341 KFB48660.1 GANO01004123 JAB55748.1 EAA10809.4 GFDL01003824 JAV31221.1 GFDF01009572 JAV04512.1 GBYB01010048 JAG79815.1 GFDL01004111 JAV30934.1 GFDL01003820 JAV31225.1 GFDL01003822 JAV31223.1 GFDF01009571 JAV04513.1 GAMC01011318 JAB95237.1 GAMC01011321 JAB95234.1 GFDL01003821 JAV31224.1 CH940648 KRF80153.1 CH902619 KPU75962.1 GBXI01015533 GBXI01010827 JAC98758.1 JAD03465.1 DS235833 EEB18122.1 GFDF01009582 JAV04502.1 CM002911 KMY92707.1 AE013599 AGB93382.1 GAMC01011320 JAB95235.1 CM000157 KRJ98588.1 CH954177 KQS70284.1 GDHF01004891 JAI47423.1 JTDY01003220 KOB69928.1 KA645887 AFP60516.1 KRF80156.1 KRF80157.1 KRF80155.1 KPU75967.1 KPU75963.1 KPU75964.1 KMY92709.1 KPU75961.1 KRF80154.1 CM000071 KRT02122.1 ACL83086.1 KRJ98592.1 KQS70292.1 AGB93381.1 KMY92705.1 OUUW01000001 SPP74076.1 AY823300 AY823301 AAF58830.4 AAV70986.1 AAV70987.1 KRJ98593.1 KQS70289.1 KMY92720.1 AF536215 AAN63887.3 BT133059 AEX93144.1 KRJ98595.1 KMY92718.1 AGB93385.1 CH963719 KRF97437.1 BT001588 AAN71343.1 KQS70291.1 ACL83085.1 KMY92710.1 GAMC01011322 JAB95233.1 KRJ98589.1 KQS70282.1 GFDL01004119 JAV30926.1 KRF80158.1 KRT02124.1 KPU75966.1 KRT02133.1 KRT02121.1 KMY92719.1 AGB93384.1 KRJ98585.1 KMY92713.1 KQS70293.1 KRF80159.1 KPU75965.1 KMY92715.1 AGB93383.1 KQS70287.1 KRJ98586.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9J2J4
A0A2A4JJ12
A0A2A4JHX1
A0A212FJ06
A0A1Q3FUE3
A0A1Q3FUT6
+ More
A0A1S4GRV7 D6W7C2 A0A1I8JU81 A7UU78 A0A084WEL6 A0A1S4GRV3 U5EU00 Q7PP97 A0A1Q3FUC3 A0A1L8DDI5 A0A182WQL0 A0A0C9Q7B4 A0A1Q3FTL0 A0A1Q3FUI8 A0A1Q3FUV2 K7IPU7 A0A1L8DDG6 W8B9C3 W8C0T6 A0A1I8NH71 A0A1Q3FUE7 A0A0Q9W4V5 A0A0P8XMR1 A0A0A1WIA3 E0VXL6 A0A1I8PAH7 A0A1L8DDH4 A0A0J9R9I6 A0A0B4KEI9 W8B271 A0A0R1DMY3 A0A0Q5WA09 A0A1W4VQP6 A0A0K8W891 A0A1W4VCN5 A0A0L7L344 T1PAX7 A0A0Q9W5H4 A0A0Q9WGP0 A0A0P8ZMI0 A0A0P8XMV7 A0A0N8P028 A0A0J9RAH7 A0A0P9AD25 A0A0Q9W508 A0A1I8NH63 A0A0R3NMI1 B7YZR3 A0A0R1DTU8 A0A0Q5WAC6 A0A0B4KF62 A0A0J9TXZ2 A0A3B0JLF9 Q5PXF9 A0A0R1DTZ7 A0A0Q5W9T9 A0A1W4VPD3 A0A0J9TY09 Q8IT87 H1UUH4 A0A0R1DU92 A0A0J9R9G6 A0A0B4KFJ4 A0A0Q9X0M2 A0A1W4VCN0 Q8IGU8 A0A0Q5W9Y9 A0A1W4VQQ1 B7YZR4 A0A0J9TXZ7 W8BE11 A0A0R1DPY4 A0A0Q5WM09 A0A1W4VQ35 A0A1Q3FTX1 A0A0Q9WGI4 A0A1I8NH75 A0A0R3NMX7 A0A0P9BWT0 A0A0R3NT21 A0A0R3NMK4 A0A0J9RAI8 A0A0B4KEP9 A0A0R1DTY9 A0A0J9R9G0 A0A0Q5WGU4 A0A0Q9WFP3 A0A1W4VQ39 A0A0P8YA19 A0A0J9TY03 A0A0B4KEH2 A0A0Q5WLK2 A0A0R1DMT9 A0A1W4VCC6
A0A1S4GRV7 D6W7C2 A0A1I8JU81 A7UU78 A0A084WEL6 A0A1S4GRV3 U5EU00 Q7PP97 A0A1Q3FUC3 A0A1L8DDI5 A0A182WQL0 A0A0C9Q7B4 A0A1Q3FTL0 A0A1Q3FUI8 A0A1Q3FUV2 K7IPU7 A0A1L8DDG6 W8B9C3 W8C0T6 A0A1I8NH71 A0A1Q3FUE7 A0A0Q9W4V5 A0A0P8XMR1 A0A0A1WIA3 E0VXL6 A0A1I8PAH7 A0A1L8DDH4 A0A0J9R9I6 A0A0B4KEI9 W8B271 A0A0R1DMY3 A0A0Q5WA09 A0A1W4VQP6 A0A0K8W891 A0A1W4VCN5 A0A0L7L344 T1PAX7 A0A0Q9W5H4 A0A0Q9WGP0 A0A0P8ZMI0 A0A0P8XMV7 A0A0N8P028 A0A0J9RAH7 A0A0P9AD25 A0A0Q9W508 A0A1I8NH63 A0A0R3NMI1 B7YZR3 A0A0R1DTU8 A0A0Q5WAC6 A0A0B4KF62 A0A0J9TXZ2 A0A3B0JLF9 Q5PXF9 A0A0R1DTZ7 A0A0Q5W9T9 A0A1W4VPD3 A0A0J9TY09 Q8IT87 H1UUH4 A0A0R1DU92 A0A0J9R9G6 A0A0B4KFJ4 A0A0Q9X0M2 A0A1W4VCN0 Q8IGU8 A0A0Q5W9Y9 A0A1W4VQQ1 B7YZR4 A0A0J9TXZ7 W8BE11 A0A0R1DPY4 A0A0Q5WM09 A0A1W4VQ35 A0A1Q3FTX1 A0A0Q9WGI4 A0A1I8NH75 A0A0R3NMX7 A0A0P9BWT0 A0A0R3NT21 A0A0R3NMK4 A0A0J9RAI8 A0A0B4KEP9 A0A0R1DTY9 A0A0J9R9G0 A0A0Q5WGU4 A0A0Q9WFP3 A0A1W4VQ39 A0A0P8YA19 A0A0J9TY03 A0A0B4KEH2 A0A0Q5WLK2 A0A0R1DMT9 A0A1W4VCC6
PDB
5VMS
E-value=5.45536e-119,
Score=1096
Ontologies
GO
PANTHER
Topology
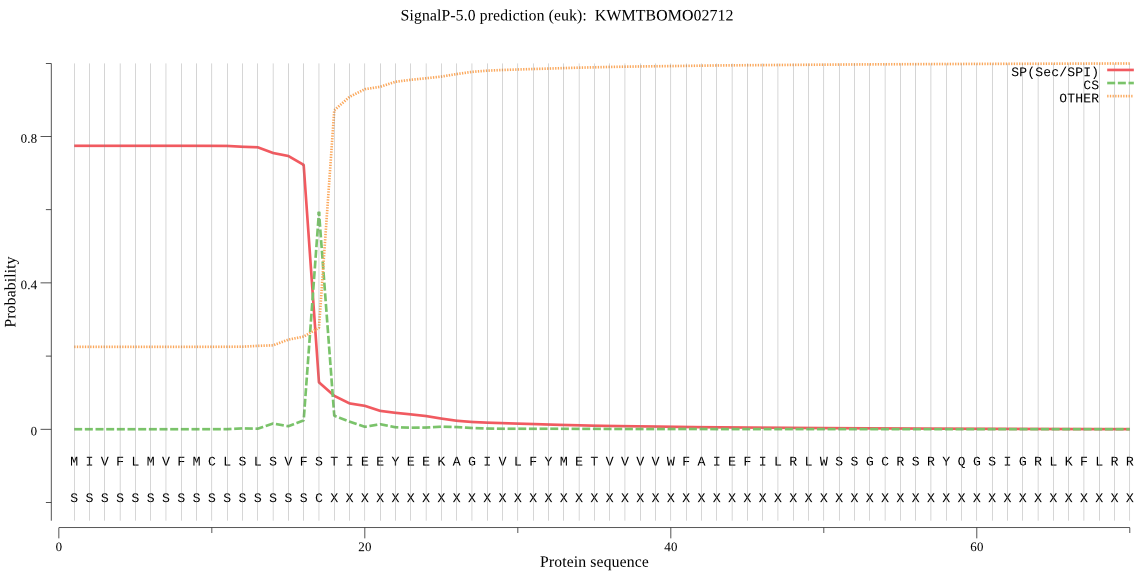
SignalP
Position: 1 - 17,
Likelihood: 0.773730
Length:
646
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
114.55493
Exp number, first 60 AAs:
38.60091
Total prob of N-in:
0.88868
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 28
TMhelix
29 - 51
inside
52 - 71
TMhelix
72 - 91
outside
92 - 136
TMhelix
137 - 156
inside
157 - 168
TMhelix
169 - 186
outside
187 - 200
TMhelix
201 - 223
inside
224 - 646

Population Genetic Test Statistics
Pi
252.942987
Theta
210.093204
Tajima's D
0.869097
CLR
0.136264
CSRT
0.623818809059547
Interpretation
Possibly Positive selection
