Gene
KWMTBOMO02703
Annotation
hypothetical_protein_RR46_11830_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.153 Extracellular Reliability : 1.357 Nuclear Reliability : 1.662
Sequence
CDS
ATGGGGATAGTAACGAGTAATCACACTAGGCGTATAAGTTTTGATAATACGTTTGCGATAAATTCGGCTTTAGCATTACAAACAACTATTAAAGGCGAAACCCCTATCGTTTTCGAAAACTTAGCCACGGAAACCGAAAATGAATTAGAATTTAAAGACAAACACATCCCCGGGAACAAACAACAAGGAGATGGAGATTATTGGTCTTGGAGAGTGGAGAACCTCAAAAAAGAGCATCAGCTAATCAATCAAATAATTGAAACCGAGTATGAAAAAAAAATTAATTCTATTAACAGTATTTGCGAACCACCTAAAGTTACTCACGAAAAATTATTGAAACTGAATCCATGCTTCCAAATGAGGACAAAGATGATTCAATGCTACACAGAAAATCCACATCAACCTTTAAAGTGTTCGGAAGCTGTTCAGGCTTTTAGTAATTGTGTTGCATACTCAAGAGCTGAAGATTAA
Protein
MGIVTSNHTRRISFDNTFAINSALALQTTIKGETPIVFENLATETENELEFKDKHIPGNKQQGDGDYWSWRVENLKKEHQLINQIIETEYEKKINSINSICEPPKVTHEKLLKLNPCFQMRTKMIQCYTENPHQPLKCSEAVQAFSNCVAYSRAED
Summary
Uniprot
EMBL
Proteomes
Pfam
PF00069 Pkinase
Interpro
ProteinModelPortal
Ontologies
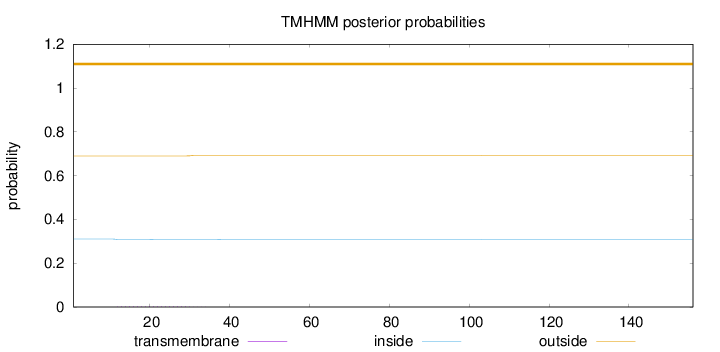
Topology
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04775
Exp number, first 60 AAs:
0.04775
Total prob of N-in:
0.31047
outside
1 - 156
Population Genetic Test Statistics
Pi
58.243602
Theta
201.536173
Tajima's D
0.249975
CLR
0.005879
CSRT
0.441477926103695
Interpretation
Uncertain
