Gene
KWMTBOMO02702 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003461
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_transmembrane_protease_serine_9_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.989
Sequence
CDS
ATGTTAGGGATTCAATTTGCGGTGCTTTGGGCTGGTCTATTCGTTATCTGTAGCGGGAATGTGCCCGAAACAGATTACGGATTTTCCTCAAAAGATGGCGCGTTTTATACTGACGATGCAATTGTGATTAATGCCCATGTAGACCGATCAAAACGTGATATAACGAATTCAACCAGAGACGGAAAACAACTTCTGCTATTGCAAGCTAGACAGCAATCAGACGAGGGCGTTTTCGGCGAGGAATGTGTATCGACAATGGGCAAGAAGGGTACATGCAAAAGTTTCCGGGATTGTTATCCACTGTTTAAAGTTGCCGATCTTTCTGGCTGGGATGGTTGGGTGATGGGACATTACGACACGTGTAGCTACATAAGCGCAGACAACATGGAGGTTTTTGGTGTCTGCTGTACAGAGCCGGTTGTTACCCCTCCTCAGCAAGAACCGGATGTGCAAAGATTAGGATTATTGCCGGGTCAACTACCTCCGATACCTGCCTTCGCAGGTCGTGTTTCTGCATTGACGGCTCAGTGGCCTTTTGAAGGTATTAGCGAGGTTCGTCAGATACCGGCCCAATGGCCGCCCACACTTCCGCCTCTACCTACCCACCCGCCTGACCACACAGCGCCAACCCATCCACCTGCAATGGGTATCACGAAACCACCACAAGCTCAACCAACACCACCCACTACTACTTGGGGAACAAAGCCGCCCAGTACCACCAAGCCTACAACGAAACAGACATGGCCTCCAGCATTCCCAACGCAGCCGAGCAAGCCATCTCAGCCTGCAGTCAGCGGCGCTTGCGGTATGAAGAATGGACCAACGGCATACGGTAGCACATACGATGTTCAAGATGAAGAGCGCATTGTTGGTGGACATAACGCAGAGCTAAACGAATGGCCCTGGATCGTAGCCCTGTTTAATGCTGGGCGTCAATTTTGCGGAGGATCTATCATTGACGACAAACACGTGATATCGGCAGCACATTGCGTAGCTCATATGACTTCCTGGGACGTCGCTCGTCTCACTGCCAGACTGGGTGACTACAACATTCGAACCAACACCGAAACATCGCACATCGAACGAAAAATTAAAAGAGTTGTCAGGCATCGAGGTTTTGACATTCGCACATTGTACAACGACATTGCGATCCTGACATTAGATCAACCAGTGACATTTACAAAGAACATTCGTCCCATTTGTTTACCGAGCGGCGGTCGGGCTTATGCTGGCCTTGTTGCTACCGTCATCGGATGGGGCAGCTTGCGAGAGAGTGGTCCGCAGCCGTCTGTTCTTCAAGAAGTATCAATTCCGATTTGGACTAACTCTGAATGTCGACTTAAATACGGCCCCGCTGCTCCGGGCGGTATAGTCGATCACATGATTTGCGCTGGAAAAGCTAGTATGGATTCCTGCAGTGGTGATAGTGGCGGGCCACTGATGGTCAACGAAGGTGGTACGTGGAACCAAGTCGGAATCGTATCATGGGGTATCGGTTGCGGTAAAGGACAGTACCCAGGTGTATACACACGCATCACTGCATTCCTACCTTGGATACAGAAGAACTCCAAGTGA
Protein
MLGIQFAVLWAGLFVICSGNVPETDYGFSSKDGAFYTDDAIVINAHVDRSKRDITNSTRDGKQLLLLQARQQSDEGVFGEECVSTMGKKGTCKSFRDCYPLFKVADLSGWDGWVMGHYDTCSYISADNMEVFGVCCTEPVVTPPQQEPDVQRLGLLPGQLPPIPAFAGRVSALTAQWPFEGISEVRQIPAQWPPTLPPLPTHPPDHTAPTHPPAMGITKPPQAQPTPPTTTWGTKPPSTTKPTTKQTWPPAFPTQPSKPSQPAVSGACGMKNGPTAYGSTYDVQDEERIVGGHNAELNEWPWIVALFNAGRQFCGGSIIDDKHVISAAHCVAHMTSWDVARLTARLGDYNIRTNTETSHIERKIKRVVRHRGFDIRTLYNDIAILTLDQPVTFTKNIRPICLPSGGRAYAGLVATVIGWGSLRESGPQPSVLQEVSIPIWTNSECRLKYGPAAPGGIVDHMICAGKASMDSCSGDSGGPLMVNEGGTWNQVGIVSWGIGCGKGQYPGVYTRITAFLPWIQKNSK
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
6O1G
E-value=4.72153e-43,
Score=440
Ontologies
GO
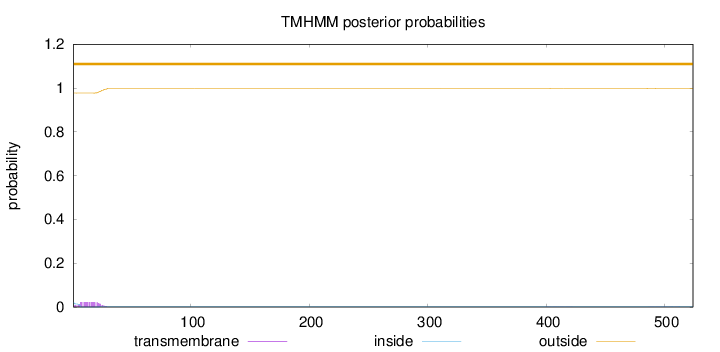
Topology
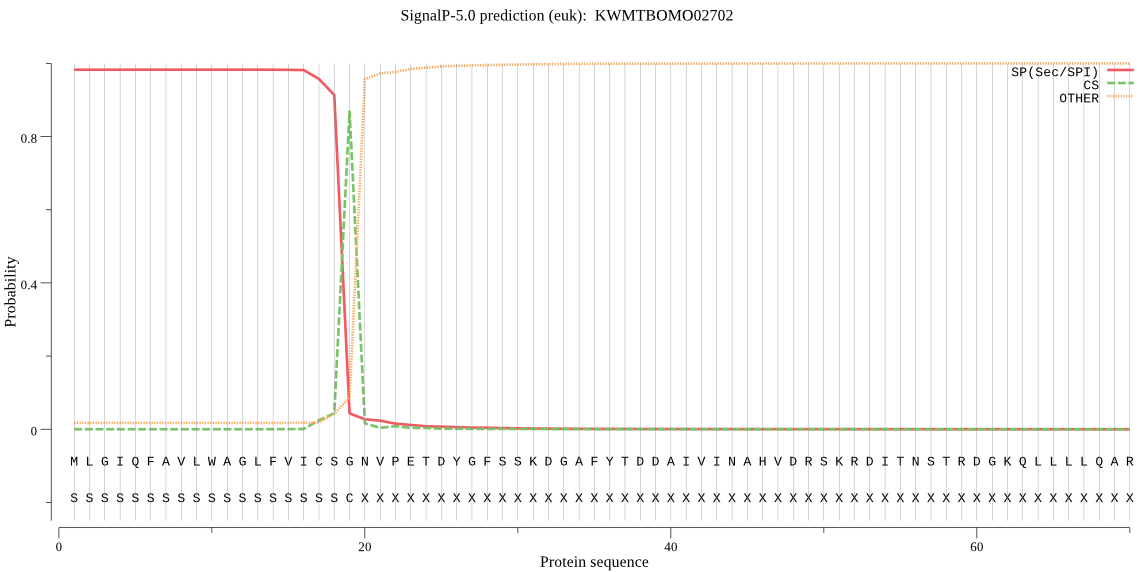
SignalP
Position: 1 - 19,
Likelihood: 0.982407
Length:
524
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48482
Exp number, first 60 AAs:
0.46475
Total prob of N-in:
0.02242
outside
1 - 524
Population Genetic Test Statistics
Pi
263.642612
Theta
188.924358
Tajima's D
1.602894
CLR
0.552294
CSRT
0.809809509524524
Interpretation
Uncertain
