Gene
KWMTBOMO02700
Pre Gene Modal
BGIBMGA003737
Annotation
PREDICTED:_phosphatidylinositol_glycan_anchor_biosynthesis_class_U_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.994
Sequence
CDS
ATGGGTGCGGTTGTAAAATATGTAATAGCCGGCTTAGTTAGAATCTTCCTTAACTGTCAAGAGAAATATTTATCAGATGTGTCTCAGGAATCAAAGGATATGGTTATGACAGAAAGTCAGTTAAAAGAGACATCTGAATATGTTCTGTCTGTCTATTTATTTAATCCATACTCTATATTGAACTGTGTTGGGATGACAACAACAGTTTTGCAGAATATGTTGTTAGCTGCAGCACTTTGTGGTGCGGCTCATGGATATGTGATAATATCCTGTGCGTTCTTAGCCTTTGCTACTCATCAAGCATTATACCCTGTCTTACTGATAGTACCAACGGCTATGATATTGGCTGAAGTGAACAAGGGATGTAATAAATGTTCATATATTAAAACATTGTTGGCCTTTGTTTTGAGTTGGAGCTTTGTGGTTTATATATCTGCATTTATAATGGATGGTTCTTATAATTATGTTTACAATACATATGGTTTTATATTGACTGTTCCAGACCTAAAACCAAATATTGGACTTTTTTGGTACTTTTTCACGGAAATGTTTGAGCACTTTAGATTACTGTTTGTATGTGCTTTCCAAATAAACGCCCTGATACTGTACATAGTACCATTGACTTTGCGATTTAAGAAGGAACCAATATTATTGGTGACTGTGCTATTAGCACTGTCAACTGTATTCAGATCTTACCCTTGTGTGGGTGATGTTGGCTTCTATATGGCATTCATACCAATGTGGAAACACCTTTTTACATTTATGCAACAAAAATTTATTGTTGCCTGTGCATTCATCATTACTTCAGCTTTGGGACCGACAGTATGGCATTTGTGGATATATTCTGGATCAGCTAATGCCAACTTTTTCTTTGGTGTCACCTTATCTTTTGCAACAGCACAAATATTCCTTATAACTGACTTATTGTTTGCGTATATTAAAAGAGAATATACTCTTAAGAATGGATCTTCGAGGACCATTGATGGAAAACCTGCCAAATTAGTTCTGCGATAA
Protein
MGAVVKYVIAGLVRIFLNCQEKYLSDVSQESKDMVMTESQLKETSEYVLSVYLFNPYSILNCVGMTTTVLQNMLLAAALCGAAHGYVIISCAFLAFATHQALYPVLLIVPTAMILAEVNKGCNKCSYIKTLLAFVLSWSFVVYISAFIMDGSYNYVYNTYGFILTVPDLKPNIGLFWYFFTEMFEHFRLLFVCAFQINALILYIVPLTLRFKKEPILLVTVLLALSTVFRSYPCVGDVGFYMAFIPMWKHLFTFMQQKFIVACAFIITSALGPTVWHLWIYSGSANANFFFGVTLSFATAQIFLITDLLFAYIKREYTLKNGSSRTIDGKPAKLVLR
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
A0A2A4K2X5
A0A1E1WG90
A0A194RL49
A0A194PNC7
A0A2H1WJ61
A0A212FB91
+ More
A0A0L7LT50 A0A067R0H3 A0A2J7RIH2 A0A1B6G5R7 A0A1B6LCH5 R4G4W1 A0A224XGS1 A0A1L8DEX0 Q16H22 A0A1Y1JY68 Q16H23 A0A023F8P8 A0A0P4WGM0 A0A151IER1 E2BG09 E2AVG3 A0A195FK75 A0A151WHN1 A0A0J7KFI7 A0A195AW42 A0A158P230 A0A0A9Z335 A0A2P8YP39 A0A146L6N0 D6WSK6 A0A195DML3 A0A1S3DKF3 A0A026VZA6 A0A182Y042 U5EXN7 A0A310SE64 A0A182SNI9 A0A0C9RCL7 A0A232EYX0 A0A084VN94 A0A182QGE7 A0A182P4Y1 A0A1B0CIH0 A0A182WU74 A0A182MSM0 A0A182W776 A0A182IY49 B0W4N5 A0A1Q3FBM5 A0A182NU95 A0A336MUN4 A0A182TSP0 A0A182UUT0 A0A182L1M4 A0A2P2I1J7 Q7Q3W6 A0A2M4AL27 A0A182RTB1 A0A182K4Z0 A0A182HUT6 A0A2S2P8V0 X1WIE1 A0A131Y6X8 A0A088AFS2 A0A1E1XCV8 A0A293N790 W5JJT0 A0A1Z5KWE1 A0A0K8TLP9 A0A131XDD7 A0A1J1IMM2 L7LSV8 R7TNP9 A0A1I8NXX8 B4LRC1 B4JBC2 E0VZ27 A0A0P5YTS1 A0A0P5UC41 A0A0P4XL44 T1J4W4 A0A0P5W2I3 A0A1L8DEU0 E9GDN1 A0A1W4WZN1 J3JXY7 B4KI55 A0A1W4VWV1 A0A1A9WFX9 Q9VLK0 A0A182F6C7 B5DH86 B4Q6V7 B7QL74 A0A1A9ZD60 A0A0P5TCB6
A0A0L7LT50 A0A067R0H3 A0A2J7RIH2 A0A1B6G5R7 A0A1B6LCH5 R4G4W1 A0A224XGS1 A0A1L8DEX0 Q16H22 A0A1Y1JY68 Q16H23 A0A023F8P8 A0A0P4WGM0 A0A151IER1 E2BG09 E2AVG3 A0A195FK75 A0A151WHN1 A0A0J7KFI7 A0A195AW42 A0A158P230 A0A0A9Z335 A0A2P8YP39 A0A146L6N0 D6WSK6 A0A195DML3 A0A1S3DKF3 A0A026VZA6 A0A182Y042 U5EXN7 A0A310SE64 A0A182SNI9 A0A0C9RCL7 A0A232EYX0 A0A084VN94 A0A182QGE7 A0A182P4Y1 A0A1B0CIH0 A0A182WU74 A0A182MSM0 A0A182W776 A0A182IY49 B0W4N5 A0A1Q3FBM5 A0A182NU95 A0A336MUN4 A0A182TSP0 A0A182UUT0 A0A182L1M4 A0A2P2I1J7 Q7Q3W6 A0A2M4AL27 A0A182RTB1 A0A182K4Z0 A0A182HUT6 A0A2S2P8V0 X1WIE1 A0A131Y6X8 A0A088AFS2 A0A1E1XCV8 A0A293N790 W5JJT0 A0A1Z5KWE1 A0A0K8TLP9 A0A131XDD7 A0A1J1IMM2 L7LSV8 R7TNP9 A0A1I8NXX8 B4LRC1 B4JBC2 E0VZ27 A0A0P5YTS1 A0A0P5UC41 A0A0P4XL44 T1J4W4 A0A0P5W2I3 A0A1L8DEU0 E9GDN1 A0A1W4WZN1 J3JXY7 B4KI55 A0A1W4VWV1 A0A1A9WFX9 Q9VLK0 A0A182F6C7 B5DH86 B4Q6V7 B7QL74 A0A1A9ZD60 A0A0P5TCB6
Pubmed
26354079
22118469
26227816
24845553
17510324
28004739
+ More
25474469 20798317 21347285 25401762 29403074 26823975 18362917 19820115 24508170 30249741 25244985 28648823 24438588 20966253 12364791 14747013 17210077 28503490 20920257 23761445 28528879 26369729 28049606 25576852 23254933 17994087 20566863 21292972 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249
25474469 20798317 21347285 25401762 29403074 26823975 18362917 19820115 24508170 30249741 25244985 28648823 24438588 20966253 12364791 14747013 17210077 28503490 20920257 23761445 28528879 26369729 28049606 25576852 23254933 17994087 20566863 21292972 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249
EMBL
NWSH01000208
PCG78399.1
GDQN01005076
JAT85978.1
KQ460207
KPJ16706.1
+ More
KQ459597 KPI94822.1 ODYU01008931 SOQ52942.1 AGBW02009362 OWR51012.1 JTDY01000155 KOB78554.1 KK852796 KDR16382.1 NEVH01003500 PNF40634.1 GECZ01029440 GECZ01021740 GECZ01012007 JAS40329.1 JAS48029.1 JAS57762.1 GEBQ01018578 JAT21399.1 ACPB03003772 GAHY01001030 JAA76480.1 GFTR01006202 JAW10224.1 GFDF01009096 JAV04988.1 CH478212 EAT33542.1 GEZM01097365 JAV54249.1 EAT33541.1 GBBI01000957 JAC17755.1 GDRN01064242 GDRN01064240 JAI64877.1 KQ977858 KYM99303.1 GL448096 EFN85411.1 GL443111 EFN62573.1 KQ981523 KYN40404.1 KQ983112 KYQ47346.1 LBMM01008210 KMQ89052.1 KQ976731 KYM76250.1 ADTU01006971 GBHO01005313 GBRD01001670 JAG38291.1 JAG64151.1 PYGN01000455 PSN46026.1 GDHC01015180 JAQ03449.1 KQ971354 EFA07117.1 KQ980724 KYN14071.1 KK107670 QOIP01000010 EZA48194.1 RLU17539.1 GANO01000862 JAB59009.1 KQ947353 OAD46869.1 GBYB01009725 GBYB01014294 JAG79492.1 JAG84061.1 NNAY01001634 OXU23368.1 ATLV01014730 KE524984 KFB39438.1 AXCN02000770 AJWK01013324 AJWK01013325 AXCM01003618 DS231838 EDS33880.1 GFDL01010069 JAV24976.1 UFQT01001254 SSX29668.1 IACF01002220 LAB67882.1 AAAB01008964 EAA12300.4 GGFK01007997 MBW41318.1 APCN01002478 GGMR01013231 MBY25850.1 ABLF02030340 GEFM01002115 JAP73681.1 GFAC01002101 JAT97087.1 GFWV01022763 MAA47490.1 ADMH02000936 ETN64632.1 GFJQ02007527 JAV99442.1 GDAI01002753 JAI14850.1 GEFH01004980 JAP63601.1 CVRI01000054 CRL00804.1 GACK01010357 JAA54677.1 AMQN01002679 KB309853 ELT93166.1 CH940649 EDW64591.1 CH916368 EDW02927.1 DS235849 EEB18633.1 GDIP01054766 JAM48949.1 GDIP01230835 GDIP01225083 GDIP01118748 GDIQ01038928 LRGB01000687 JAL84966.1 JAN55809.1 KZS16662.1 GDIP01240338 JAI83063.1 JH431850 GDIP01091937 JAM11778.1 GFDF01009118 JAV04966.1 GL732540 EFX82447.1 BT128109 AEE63070.1 CH933807 EDW12348.1 AE014134 AY058668 AAF52689.1 AAL13897.1 CH379058 EDY69661.1 CM000361 CM002910 EDX04266.1 KMY89119.1 ABJB010755356 DS963775 EEC19596.1 GDIP01134108 JAL69606.1
KQ459597 KPI94822.1 ODYU01008931 SOQ52942.1 AGBW02009362 OWR51012.1 JTDY01000155 KOB78554.1 KK852796 KDR16382.1 NEVH01003500 PNF40634.1 GECZ01029440 GECZ01021740 GECZ01012007 JAS40329.1 JAS48029.1 JAS57762.1 GEBQ01018578 JAT21399.1 ACPB03003772 GAHY01001030 JAA76480.1 GFTR01006202 JAW10224.1 GFDF01009096 JAV04988.1 CH478212 EAT33542.1 GEZM01097365 JAV54249.1 EAT33541.1 GBBI01000957 JAC17755.1 GDRN01064242 GDRN01064240 JAI64877.1 KQ977858 KYM99303.1 GL448096 EFN85411.1 GL443111 EFN62573.1 KQ981523 KYN40404.1 KQ983112 KYQ47346.1 LBMM01008210 KMQ89052.1 KQ976731 KYM76250.1 ADTU01006971 GBHO01005313 GBRD01001670 JAG38291.1 JAG64151.1 PYGN01000455 PSN46026.1 GDHC01015180 JAQ03449.1 KQ971354 EFA07117.1 KQ980724 KYN14071.1 KK107670 QOIP01000010 EZA48194.1 RLU17539.1 GANO01000862 JAB59009.1 KQ947353 OAD46869.1 GBYB01009725 GBYB01014294 JAG79492.1 JAG84061.1 NNAY01001634 OXU23368.1 ATLV01014730 KE524984 KFB39438.1 AXCN02000770 AJWK01013324 AJWK01013325 AXCM01003618 DS231838 EDS33880.1 GFDL01010069 JAV24976.1 UFQT01001254 SSX29668.1 IACF01002220 LAB67882.1 AAAB01008964 EAA12300.4 GGFK01007997 MBW41318.1 APCN01002478 GGMR01013231 MBY25850.1 ABLF02030340 GEFM01002115 JAP73681.1 GFAC01002101 JAT97087.1 GFWV01022763 MAA47490.1 ADMH02000936 ETN64632.1 GFJQ02007527 JAV99442.1 GDAI01002753 JAI14850.1 GEFH01004980 JAP63601.1 CVRI01000054 CRL00804.1 GACK01010357 JAA54677.1 AMQN01002679 KB309853 ELT93166.1 CH940649 EDW64591.1 CH916368 EDW02927.1 DS235849 EEB18633.1 GDIP01054766 JAM48949.1 GDIP01230835 GDIP01225083 GDIP01118748 GDIQ01038928 LRGB01000687 JAL84966.1 JAN55809.1 KZS16662.1 GDIP01240338 JAI83063.1 JH431850 GDIP01091937 JAM11778.1 GFDF01009118 JAV04966.1 GL732540 EFX82447.1 BT128109 AEE63070.1 CH933807 EDW12348.1 AE014134 AY058668 AAF52689.1 AAL13897.1 CH379058 EDY69661.1 CM000361 CM002910 EDX04266.1 KMY89119.1 ABJB010755356 DS963775 EEC19596.1 GDIP01134108 JAL69606.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000027135
+ More
UP000235965 UP000015103 UP000008820 UP000078542 UP000008237 UP000000311 UP000078541 UP000075809 UP000036403 UP000078540 UP000005205 UP000245037 UP000007266 UP000078492 UP000079169 UP000053097 UP000279307 UP000076408 UP000075901 UP000215335 UP000030765 UP000075886 UP000075885 UP000092461 UP000076407 UP000075883 UP000075920 UP000075880 UP000002320 UP000075884 UP000075902 UP000075903 UP000075882 UP000007062 UP000075900 UP000075881 UP000075840 UP000007819 UP000005203 UP000000673 UP000183832 UP000014760 UP000095300 UP000008792 UP000001070 UP000009046 UP000076858 UP000000305 UP000192223 UP000009192 UP000192221 UP000091820 UP000000803 UP000069272 UP000001819 UP000000304 UP000001555 UP000092445
UP000235965 UP000015103 UP000008820 UP000078542 UP000008237 UP000000311 UP000078541 UP000075809 UP000036403 UP000078540 UP000005205 UP000245037 UP000007266 UP000078492 UP000079169 UP000053097 UP000279307 UP000076408 UP000075901 UP000215335 UP000030765 UP000075886 UP000075885 UP000092461 UP000076407 UP000075883 UP000075920 UP000075880 UP000002320 UP000075884 UP000075902 UP000075903 UP000075882 UP000007062 UP000075900 UP000075881 UP000075840 UP000007819 UP000005203 UP000000673 UP000183832 UP000014760 UP000095300 UP000008792 UP000001070 UP000009046 UP000076858 UP000000305 UP000192223 UP000009192 UP000192221 UP000091820 UP000000803 UP000069272 UP000001819 UP000000304 UP000001555 UP000092445
Pfam
Interpro
IPR009600
PIG-U
+ More
IPR019176 Cytochrome_B561-rel
IPR032384 Kif23_Arf-bd
IPR038105 Kif23_Arf-bd_sf
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR017927 FAD-bd_FR_type
IPR013112 FAD-bd_8
IPR017938 Riboflavin_synthase-like_b-brl
IPR029648 NOX5
IPR039261 FNR_nucleotide-bd
IPR013121 Fe_red_NAD-bd_6
IPR013130 Fe3_Rdtase_TM_dom
IPR019176 Cytochrome_B561-rel
IPR032384 Kif23_Arf-bd
IPR038105 Kif23_Arf-bd_sf
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR017927 FAD-bd_FR_type
IPR013112 FAD-bd_8
IPR017938 Riboflavin_synthase-like_b-brl
IPR029648 NOX5
IPR039261 FNR_nucleotide-bd
IPR013121 Fe_red_NAD-bd_6
IPR013130 Fe3_Rdtase_TM_dom
Gene 3D
ProteinModelPortal
A0A2A4K2X5
A0A1E1WG90
A0A194RL49
A0A194PNC7
A0A2H1WJ61
A0A212FB91
+ More
A0A0L7LT50 A0A067R0H3 A0A2J7RIH2 A0A1B6G5R7 A0A1B6LCH5 R4G4W1 A0A224XGS1 A0A1L8DEX0 Q16H22 A0A1Y1JY68 Q16H23 A0A023F8P8 A0A0P4WGM0 A0A151IER1 E2BG09 E2AVG3 A0A195FK75 A0A151WHN1 A0A0J7KFI7 A0A195AW42 A0A158P230 A0A0A9Z335 A0A2P8YP39 A0A146L6N0 D6WSK6 A0A195DML3 A0A1S3DKF3 A0A026VZA6 A0A182Y042 U5EXN7 A0A310SE64 A0A182SNI9 A0A0C9RCL7 A0A232EYX0 A0A084VN94 A0A182QGE7 A0A182P4Y1 A0A1B0CIH0 A0A182WU74 A0A182MSM0 A0A182W776 A0A182IY49 B0W4N5 A0A1Q3FBM5 A0A182NU95 A0A336MUN4 A0A182TSP0 A0A182UUT0 A0A182L1M4 A0A2P2I1J7 Q7Q3W6 A0A2M4AL27 A0A182RTB1 A0A182K4Z0 A0A182HUT6 A0A2S2P8V0 X1WIE1 A0A131Y6X8 A0A088AFS2 A0A1E1XCV8 A0A293N790 W5JJT0 A0A1Z5KWE1 A0A0K8TLP9 A0A131XDD7 A0A1J1IMM2 L7LSV8 R7TNP9 A0A1I8NXX8 B4LRC1 B4JBC2 E0VZ27 A0A0P5YTS1 A0A0P5UC41 A0A0P4XL44 T1J4W4 A0A0P5W2I3 A0A1L8DEU0 E9GDN1 A0A1W4WZN1 J3JXY7 B4KI55 A0A1W4VWV1 A0A1A9WFX9 Q9VLK0 A0A182F6C7 B5DH86 B4Q6V7 B7QL74 A0A1A9ZD60 A0A0P5TCB6
A0A0L7LT50 A0A067R0H3 A0A2J7RIH2 A0A1B6G5R7 A0A1B6LCH5 R4G4W1 A0A224XGS1 A0A1L8DEX0 Q16H22 A0A1Y1JY68 Q16H23 A0A023F8P8 A0A0P4WGM0 A0A151IER1 E2BG09 E2AVG3 A0A195FK75 A0A151WHN1 A0A0J7KFI7 A0A195AW42 A0A158P230 A0A0A9Z335 A0A2P8YP39 A0A146L6N0 D6WSK6 A0A195DML3 A0A1S3DKF3 A0A026VZA6 A0A182Y042 U5EXN7 A0A310SE64 A0A182SNI9 A0A0C9RCL7 A0A232EYX0 A0A084VN94 A0A182QGE7 A0A182P4Y1 A0A1B0CIH0 A0A182WU74 A0A182MSM0 A0A182W776 A0A182IY49 B0W4N5 A0A1Q3FBM5 A0A182NU95 A0A336MUN4 A0A182TSP0 A0A182UUT0 A0A182L1M4 A0A2P2I1J7 Q7Q3W6 A0A2M4AL27 A0A182RTB1 A0A182K4Z0 A0A182HUT6 A0A2S2P8V0 X1WIE1 A0A131Y6X8 A0A088AFS2 A0A1E1XCV8 A0A293N790 W5JJT0 A0A1Z5KWE1 A0A0K8TLP9 A0A131XDD7 A0A1J1IMM2 L7LSV8 R7TNP9 A0A1I8NXX8 B4LRC1 B4JBC2 E0VZ27 A0A0P5YTS1 A0A0P5UC41 A0A0P4XL44 T1J4W4 A0A0P5W2I3 A0A1L8DEU0 E9GDN1 A0A1W4WZN1 J3JXY7 B4KI55 A0A1W4VWV1 A0A1A9WFX9 Q9VLK0 A0A182F6C7 B5DH86 B4Q6V7 B7QL74 A0A1A9ZD60 A0A0P5TCB6
Ontologies
PATHWAY
GO
PANTHER
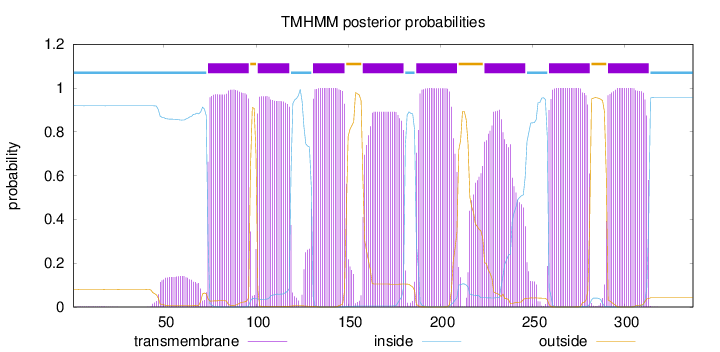
Topology
Length:
337
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
172.58771
Exp number, first 60 AAs:
1.89496
Total prob of N-in:
0.92001
inside
1 - 73
TMhelix
74 - 96
outside
97 - 100
TMhelix
101 - 118
inside
119 - 130
TMhelix
131 - 148
outside
149 - 157
TMhelix
158 - 180
inside
181 - 186
TMhelix
187 - 209
outside
210 - 223
TMhelix
224 - 246
inside
247 - 258
TMhelix
259 - 281
outside
282 - 290
TMhelix
291 - 313
inside
314 - 337
Population Genetic Test Statistics
Pi
279.22224
Theta
211.336913
Tajima's D
1.011302
CLR
0.361859
CSRT
0.665366731663417
Interpretation
Uncertain
