Pre Gene Modal
BGIBMGA003738
Annotation
PREDICTED:_uncharacterized_protein_LOC101741587_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.507
Sequence
CDS
ATGAAGTATTTCCTTTACGCAGTGACATTCGTCTTATCAGAATGTGCTATTTTGCCCAGTATACCAACACATCTACTTCCATGTTATCGCAATGGTGCCCCAGAATTAGTTGCACCCCGGCGTCTTGATGTTTTTTTGTCTTTGTTGAGAAAAATCGAGATACACACGCAGCTAGACATGAGGATGATTACTCCAGCTTTACTAAGGAGTCTTCGTTTAGATGGTATAGAACAGCTTTCCGCAACAGCGGTTGAAACGGAATTCGTGCTGCCATTTAGAGCATCTGCTTTCCAATTTCACAAATACAAACTTTTGATGGATTTATTCTTGCCTAGTCAGAACCTAGTGGATGTTGATGAAATATTAACCGTACCAGAAAAATGTTTACTACATCGGATGCTAAGTTCAGCGGTGCGACTATGGGAAAGAGGCGATGAGAACGTTGCGTGTCCTCTGACATCGGAACAATCGCAAAACATGATATCACAAAGTAATACTAGTATCATCAGCAGATGTCCAATCGAAGATGGAGTAATACAAACTGAATGGGGAACTATATCTCCTGGGACAGTCATGGCTGCTGTAGCCTCAGCTCTTGAAAATCAACGAGTATCGATGAGCGACATTTTCAACGCTAATATTTTTAAGGAAGACGTAGCTGAACCTCTAATTAATTTAGCTTTACAAGACTGGTATGAAAACATAGAAACTCTTGATACAAATAATCAAAAACAGCTCGAGAATGTTGATATAAACAATATTTGGGTAGCCACACTAGGAGGTGATTTGGCTGAAGTAGTAGTTAACCAAGGTCCTAGAGTGGGTGCCTCTTCGCAAAGAATGAACATAGGCACCAACAATAGATGGAACGATACGCTACTTCCTCGTGCGCTATACATTTTTCCTCAGAATGCTTCAATCATTGATTGGCACATCACTGATGCAGAAATATTAGCTGGAATCGACGGTTTGATTTTGTCAAGATACATTTCGAAATGGGTAGAACAAAGAAGATCTTTAAGATTGTCTCAAGTAATAGACATGTACTACTCCAACGAAGGTGTTTCATTTGATCCTAACGTTAAGGCTTGTAATAGATTGTCCCTATTCACTGATGTCTTTGATAGTTCTGATTTAAAGACGGAGGTTGCTAGGTTTGCTTCTGTGTTATCTCTGAGGCAAATAACTGTGTATATACCACAGGATGAGATGGAACGAATCACCGAAGCCGCTGTGACTGCATTCACAGATTATTTGTCGTCAATCTTGCGACAGAATCAAAGAGAGTGCCGCATGGGCCGTACTATACCTGTAATGGACTTGGTCGTAGCAACTGACAGCTCGTGGGGACGTTACGAAGTAGAACAGTTTTTGTCGTGGGTCAGTGGAGCTCTAGAGATGAATCTGATGAAGAGTACGATATCATTATTGCACGGAAACACAGGGGCGTGGATTGCACCGCCATCAAGTAATTTAACGGCACTGTTTTCACACGTACAAAATTTCACAGATACGTGGCCAAATCGTTTGAATCTGCCAAACATAATATCGACTGCCATTCGGTATTCACTCAATAGGACTTTAAATGAAATAGAGAGTCAGGCTAGTGCTGGAGTCAGTACTGTTGTGCTCGTTATTAGTCCGAGTGATCGACCGTCCAGTACGGAGATGGAGAGAGCCCGGAGCCTCATGAGTACACTGAGAAATAGCTTCTTTGACGTATACTTTTCTTACGTCGCGCGGGACGTGACTGATTTTCAGAACATCAACAATGAATATTTAGATTATTCTGAGCTGTTCGTTACGACGACATCAACTTCAGTGAGTGACGTAATCAACGGGGTGGACACGTACGTAGTGCAAAATAGTATACCGATGAGGTTAATGGGGGCTCAGTGTCAAGTTAACGATACCATATTTGAACAAGTTGAATATGAAGACTTCGTCTTACCGAATCGGCAAACACATTACAGAATACATCCTTTTTATTTGAGACAACAAGCCCTGGTTCAAGTTCAGTTTCGTAACGACGGCCAAGGTCGACTATTGATATGTACATGGCGCGGAGCTGACGCGTCTCACGACTGTCGTACATTATCGGAACGTGAACTGTTCACGTTCAATCTTACCACGCCATGTCCGTCACCGGAATTTTGTCCTCCAGCTCATTTTATATCAACAGCAATAAACTCTTCTAATTTATGTGCAAATAACGACTGTCGTCTTCCACATCAAGTCGGTTACTACGTCCGACATTCTGGGCTTCGCTGTTTGCCGCTTACAGGATCTTCTAATAAGAACGAGATCAGGATGTTGTTCATTTATATAATAATTTTAGGATTTTTATTAAAATAA
Protein
MKYFLYAVTFVLSECAILPSIPTHLLPCYRNGAPELVAPRRLDVFLSLLRKIEIHTQLDMRMITPALLRSLRLDGIEQLSATAVETEFVLPFRASAFQFHKYKLLMDLFLPSQNLVDVDEILTVPEKCLLHRMLSSAVRLWERGDENVACPLTSEQSQNMISQSNTSIISRCPIEDGVIQTEWGTISPGTVMAAVASALENQRVSMSDIFNANIFKEDVAEPLINLALQDWYENIETLDTNNQKQLENVDINNIWVATLGGDLAEVVVNQGPRVGASSQRMNIGTNNRWNDTLLPRALYIFPQNASIIDWHITDAEILAGIDGLILSRYISKWVEQRRSLRLSQVIDMYYSNEGVSFDPNVKACNRLSLFTDVFDSSDLKTEVARFASVLSLRQITVYIPQDEMERITEAAVTAFTDYLSSILRQNQRECRMGRTIPVMDLVVATDSSWGRYEVEQFLSWVSGALEMNLMKSTISLLHGNTGAWIAPPSSNLTALFSHVQNFTDTWPNRLNLPNIISTAIRYSLNRTLNEIESQASAGVSTVVLVISPSDRPSSTEMERARSLMSTLRNSFFDVYFSYVARDVTDFQNINNEYLDYSELFVTTTSTSVSDVINGVDTYVVQNSIPMRLMGAQCQVNDTIFEQVEYEDFVLPNRQTHYRIHPFYLRQQALVQVQFRNDGQGRLLICTWRGADASHDCRTLSERELFTFNLTTPCPSPEFCPPAHFISTAINSSNLCANNDCRLPHQVGYYVRHSGLRCLPLTGSSNKNEIRMLFIYIIILGFLLK
Summary
Similarity
Belongs to the STXBP/unc-18/SEC1 family.
Uniprot
A0A2A4K3A1
A0A1E1WAR7
A0A2H1WJ61
A0A194PN13
A0A212FB91
A0A194RHA8
+ More
A0A1L8D9I3 W5JTU0 A0A182GEU0 A0A182FPL6 Q16IE0 Q16NE4 A0A1B0D7R4 A0A182N3A2 A0A182R0D4 A0A1B6CHY1 A0A2J7R0E2 A0A182Y726 V5G1Z3 A0A1J1J8Q8 A0A182KK82 A0A182W1P6 K7JQ83 A0A1S4H6H0 A0A182HY27 A0A0A9XAX3 T1HV47 A0A3L8D4I8 A0A232EYI9 A0A1B6D0F0 A0A0L7QUI9 A0A0C9Q577 A0A088A0Z6 A0A0A1XL35 A0A088A221 A0A2A3EMU3 A0A336MV53 A0A2A3ENG3 A0A336MP36 A0A1B6F181 A0A2A3ELJ7 A0A026VXQ2 A0A154PRG2 W8BJC0 A0A1A9Z5D9 B3MCE5 A0A1L8D9X5 A0A1L8D9M0 A0A0M3QVR1 B4PC03 A0A0R1DZL7 Q8MLN4 Q6NP37 B4QD68 A0A0J9RKN8 A0A1B0G4K1 A0A1W4V5T1 A0A1L8EC68 B4KSS8 A0A0Q9W2F4 B4LNT2 Q28Z84 A0A1L8ECG4 A0A1B6LID2 B4GI45 A0A3B0IY76 B4JVJ1 A0A1Y1NED5 A0A1A9VVM6 B3NRB0 A0A0M8ZWE0 A0A1A9W7T5 B4MPL3 B4IHD6 A0A0J9RM94 A0A026VV20 A0A1W4XGK8 U5ESY0 F4WM87 Q0E8V8 A0A0R1DU80 A0A1A9XHF3 A0A0R1DXJ9 A0A034VDF0
A0A1L8D9I3 W5JTU0 A0A182GEU0 A0A182FPL6 Q16IE0 Q16NE4 A0A1B0D7R4 A0A182N3A2 A0A182R0D4 A0A1B6CHY1 A0A2J7R0E2 A0A182Y726 V5G1Z3 A0A1J1J8Q8 A0A182KK82 A0A182W1P6 K7JQ83 A0A1S4H6H0 A0A182HY27 A0A0A9XAX3 T1HV47 A0A3L8D4I8 A0A232EYI9 A0A1B6D0F0 A0A0L7QUI9 A0A0C9Q577 A0A088A0Z6 A0A0A1XL35 A0A088A221 A0A2A3EMU3 A0A336MV53 A0A2A3ENG3 A0A336MP36 A0A1B6F181 A0A2A3ELJ7 A0A026VXQ2 A0A154PRG2 W8BJC0 A0A1A9Z5D9 B3MCE5 A0A1L8D9X5 A0A1L8D9M0 A0A0M3QVR1 B4PC03 A0A0R1DZL7 Q8MLN4 Q6NP37 B4QD68 A0A0J9RKN8 A0A1B0G4K1 A0A1W4V5T1 A0A1L8EC68 B4KSS8 A0A0Q9W2F4 B4LNT2 Q28Z84 A0A1L8ECG4 A0A1B6LID2 B4GI45 A0A3B0IY76 B4JVJ1 A0A1Y1NED5 A0A1A9VVM6 B3NRB0 A0A0M8ZWE0 A0A1A9W7T5 B4MPL3 B4IHD6 A0A0J9RM94 A0A026VV20 A0A1W4XGK8 U5ESY0 F4WM87 Q0E8V8 A0A0R1DU80 A0A1A9XHF3 A0A0R1DXJ9 A0A034VDF0
Pubmed
26354079
22118469
20920257
23761445
26483478
17510324
+ More
25244985 20966253 20075255 12364791 25401762 30249741 28648823 25830018 24508170 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 28004739 21719571 25348373
25244985 20966253 20075255 12364791 25401762 30249741 28648823 25830018 24508170 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 28004739 21719571 25348373
EMBL
NWSH01000208
PCG78398.1
GDQN01007103
JAT83951.1
ODYU01008931
SOQ52942.1
+ More
KQ459597 KPI94821.1 AGBW02009362 OWR51012.1 KQ460207 KPJ16705.1 GFDF01010948 JAV03136.1 ADMH02000117 ETN67777.1 JXUM01058543 KQ562007 KXJ76930.1 CH478086 EAT34026.1 CH477828 EAT35855.1 AJVK01027142 AXCN02001120 GEDC01024222 JAS13076.1 NEVH01008258 PNF34286.1 GALX01004446 JAB64020.1 CVRI01000070 CRL07297.1 AAZX01003923 AAAB01008966 APCN01004484 GBHO01027641 JAG15963.1 ACPB03004465 QOIP01000013 RLU15417.1 NNAY01001572 OXU23556.1 GEDC01030956 GEDC01018116 JAS06342.1 JAS19182.1 KQ414735 KOC62308.1 GBYB01009208 JAG78975.1 GBXI01003044 JAD11248.1 KZ288215 PBC32582.1 UFQT01003040 SSX34484.1 PBC32581.1 UFQT01001909 SSX32132.1 GECZ01025851 JAS43918.1 PBC32580.1 KK107796 EZA47609.1 KQ435078 KZC14463.1 GAMC01016857 GAMC01016856 JAB89699.1 CH902619 EDV37269.1 GFDF01010919 JAV03165.1 GFDF01010918 JAV03166.1 CP012524 ALC42833.1 CM000158 EDW92657.1 KRK00630.1 KRK00628.1 AE013599 AAM68331.2 AGB93709.1 AGB93710.1 BT011094 AAR82760.1 CM000362 EDX08664.1 CM002911 KMY96533.1 CCAG010000826 GFDG01002462 JAV16337.1 CH933808 EDW10577.2 CH940648 KRF79283.1 EDW60152.2 CM000071 EAL25730.4 GFDG01002463 JAV16336.1 GEBQ01016579 JAT23398.1 CH479183 EDW36165.1 OUUW01000001 SPP72657.1 CH916375 EDV98459.1 GEZM01007311 JAV95250.1 CH954179 EDV57129.1 KQ435821 KOX72475.1 CH963849 EDW74052.1 CH480838 EDW49312.1 KMY96534.1 EZA47608.1 RLU15414.1 GANO01004630 JAB55241.1 GL888217 EGI64731.1 BT083392 ABI31116.2 ACQ66053.1 KRK00629.1 KRK00627.1 GAKP01018418 JAC40534.1
KQ459597 KPI94821.1 AGBW02009362 OWR51012.1 KQ460207 KPJ16705.1 GFDF01010948 JAV03136.1 ADMH02000117 ETN67777.1 JXUM01058543 KQ562007 KXJ76930.1 CH478086 EAT34026.1 CH477828 EAT35855.1 AJVK01027142 AXCN02001120 GEDC01024222 JAS13076.1 NEVH01008258 PNF34286.1 GALX01004446 JAB64020.1 CVRI01000070 CRL07297.1 AAZX01003923 AAAB01008966 APCN01004484 GBHO01027641 JAG15963.1 ACPB03004465 QOIP01000013 RLU15417.1 NNAY01001572 OXU23556.1 GEDC01030956 GEDC01018116 JAS06342.1 JAS19182.1 KQ414735 KOC62308.1 GBYB01009208 JAG78975.1 GBXI01003044 JAD11248.1 KZ288215 PBC32582.1 UFQT01003040 SSX34484.1 PBC32581.1 UFQT01001909 SSX32132.1 GECZ01025851 JAS43918.1 PBC32580.1 KK107796 EZA47609.1 KQ435078 KZC14463.1 GAMC01016857 GAMC01016856 JAB89699.1 CH902619 EDV37269.1 GFDF01010919 JAV03165.1 GFDF01010918 JAV03166.1 CP012524 ALC42833.1 CM000158 EDW92657.1 KRK00630.1 KRK00628.1 AE013599 AAM68331.2 AGB93709.1 AGB93710.1 BT011094 AAR82760.1 CM000362 EDX08664.1 CM002911 KMY96533.1 CCAG010000826 GFDG01002462 JAV16337.1 CH933808 EDW10577.2 CH940648 KRF79283.1 EDW60152.2 CM000071 EAL25730.4 GFDG01002463 JAV16336.1 GEBQ01016579 JAT23398.1 CH479183 EDW36165.1 OUUW01000001 SPP72657.1 CH916375 EDV98459.1 GEZM01007311 JAV95250.1 CH954179 EDV57129.1 KQ435821 KOX72475.1 CH963849 EDW74052.1 CH480838 EDW49312.1 KMY96534.1 EZA47608.1 RLU15414.1 GANO01004630 JAB55241.1 GL888217 EGI64731.1 BT083392 ABI31116.2 ACQ66053.1 KRK00629.1 KRK00627.1 GAKP01018418 JAC40534.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000000673
UP000069940
+ More
UP000249989 UP000069272 UP000008820 UP000092462 UP000075884 UP000075886 UP000235965 UP000076408 UP000183832 UP000075882 UP000075920 UP000002358 UP000075840 UP000015103 UP000279307 UP000215335 UP000053825 UP000005203 UP000242457 UP000053097 UP000076502 UP000092445 UP000007801 UP000092553 UP000002282 UP000000803 UP000000304 UP000092444 UP000192221 UP000009192 UP000008792 UP000001819 UP000008744 UP000268350 UP000001070 UP000078200 UP000008711 UP000053105 UP000091820 UP000007798 UP000001292 UP000192223 UP000007755 UP000092443
UP000249989 UP000069272 UP000008820 UP000092462 UP000075884 UP000075886 UP000235965 UP000076408 UP000183832 UP000075882 UP000075920 UP000002358 UP000075840 UP000015103 UP000279307 UP000215335 UP000053825 UP000005203 UP000242457 UP000053097 UP000076502 UP000092445 UP000007801 UP000092553 UP000002282 UP000000803 UP000000304 UP000092444 UP000192221 UP000009192 UP000008792 UP000001819 UP000008744 UP000268350 UP000001070 UP000078200 UP000008711 UP000053105 UP000091820 UP000007798 UP000001292 UP000192223 UP000007755 UP000092443
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K3A1
A0A1E1WAR7
A0A2H1WJ61
A0A194PN13
A0A212FB91
A0A194RHA8
+ More
A0A1L8D9I3 W5JTU0 A0A182GEU0 A0A182FPL6 Q16IE0 Q16NE4 A0A1B0D7R4 A0A182N3A2 A0A182R0D4 A0A1B6CHY1 A0A2J7R0E2 A0A182Y726 V5G1Z3 A0A1J1J8Q8 A0A182KK82 A0A182W1P6 K7JQ83 A0A1S4H6H0 A0A182HY27 A0A0A9XAX3 T1HV47 A0A3L8D4I8 A0A232EYI9 A0A1B6D0F0 A0A0L7QUI9 A0A0C9Q577 A0A088A0Z6 A0A0A1XL35 A0A088A221 A0A2A3EMU3 A0A336MV53 A0A2A3ENG3 A0A336MP36 A0A1B6F181 A0A2A3ELJ7 A0A026VXQ2 A0A154PRG2 W8BJC0 A0A1A9Z5D9 B3MCE5 A0A1L8D9X5 A0A1L8D9M0 A0A0M3QVR1 B4PC03 A0A0R1DZL7 Q8MLN4 Q6NP37 B4QD68 A0A0J9RKN8 A0A1B0G4K1 A0A1W4V5T1 A0A1L8EC68 B4KSS8 A0A0Q9W2F4 B4LNT2 Q28Z84 A0A1L8ECG4 A0A1B6LID2 B4GI45 A0A3B0IY76 B4JVJ1 A0A1Y1NED5 A0A1A9VVM6 B3NRB0 A0A0M8ZWE0 A0A1A9W7T5 B4MPL3 B4IHD6 A0A0J9RM94 A0A026VV20 A0A1W4XGK8 U5ESY0 F4WM87 Q0E8V8 A0A0R1DU80 A0A1A9XHF3 A0A0R1DXJ9 A0A034VDF0
A0A1L8D9I3 W5JTU0 A0A182GEU0 A0A182FPL6 Q16IE0 Q16NE4 A0A1B0D7R4 A0A182N3A2 A0A182R0D4 A0A1B6CHY1 A0A2J7R0E2 A0A182Y726 V5G1Z3 A0A1J1J8Q8 A0A182KK82 A0A182W1P6 K7JQ83 A0A1S4H6H0 A0A182HY27 A0A0A9XAX3 T1HV47 A0A3L8D4I8 A0A232EYI9 A0A1B6D0F0 A0A0L7QUI9 A0A0C9Q577 A0A088A0Z6 A0A0A1XL35 A0A088A221 A0A2A3EMU3 A0A336MV53 A0A2A3ENG3 A0A336MP36 A0A1B6F181 A0A2A3ELJ7 A0A026VXQ2 A0A154PRG2 W8BJC0 A0A1A9Z5D9 B3MCE5 A0A1L8D9X5 A0A1L8D9M0 A0A0M3QVR1 B4PC03 A0A0R1DZL7 Q8MLN4 Q6NP37 B4QD68 A0A0J9RKN8 A0A1B0G4K1 A0A1W4V5T1 A0A1L8EC68 B4KSS8 A0A0Q9W2F4 B4LNT2 Q28Z84 A0A1L8ECG4 A0A1B6LID2 B4GI45 A0A3B0IY76 B4JVJ1 A0A1Y1NED5 A0A1A9VVM6 B3NRB0 A0A0M8ZWE0 A0A1A9W7T5 B4MPL3 B4IHD6 A0A0J9RM94 A0A026VV20 A0A1W4XGK8 U5ESY0 F4WM87 Q0E8V8 A0A0R1DU80 A0A1A9XHF3 A0A0R1DXJ9 A0A034VDF0
Ontologies
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.881095
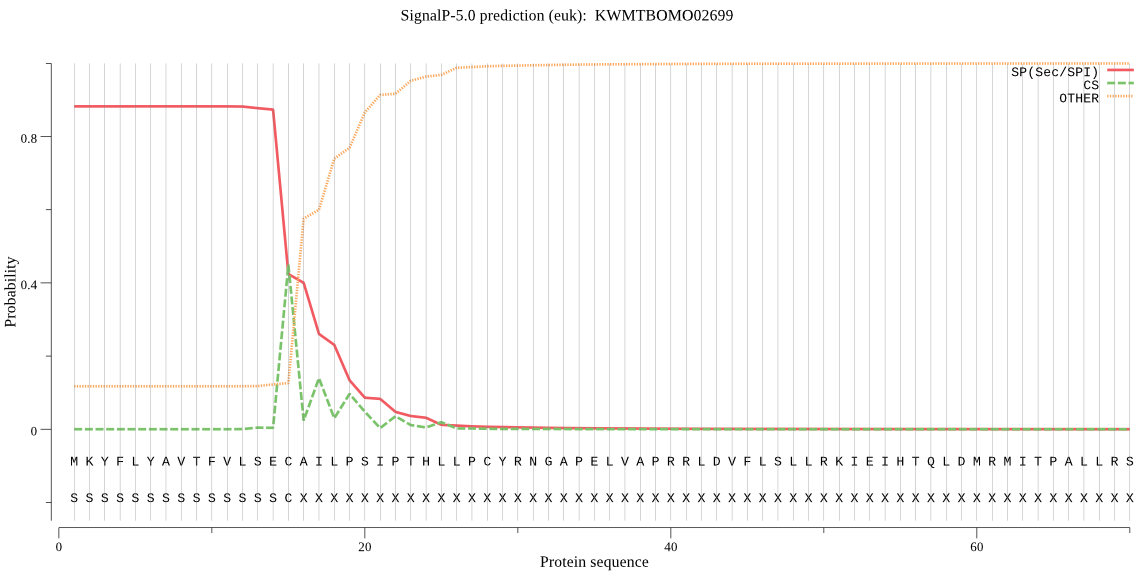
Length:
784
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.65255
Exp number, first 60 AAs:
1.06339
Total prob of N-in:
0.04997
outside
1 - 784
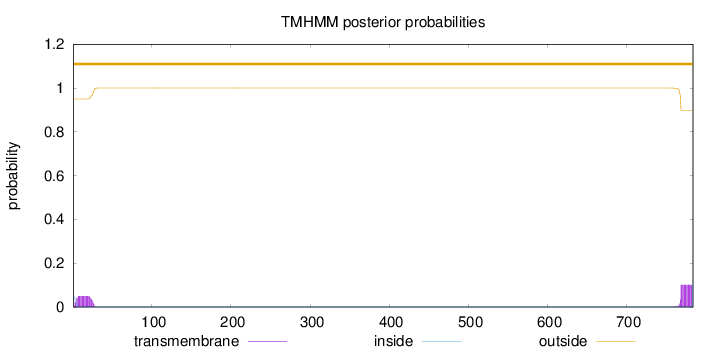
Population Genetic Test Statistics
Pi
335.442242
Theta
220.321706
Tajima's D
1.664655
CLR
0.255205
CSRT
0.818109094545273
Interpretation
Uncertain
