Gene
KWMTBOMO02696 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003458
Annotation
PREDICTED:_uncharacterized_protein_LOC106100834_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 3.33
Sequence
CDS
ATGTATTGGCTACCATTCTTTATGGTAGCTGTTGTGGGTACTACCCACGGACAACTAACGGCGCAGTCCAGTATCGCACCAGAGCTACGGGAATGCTATTTTGATCCCATACTGTTAAACAGAAACAATCTCCCTCCGACCACAATTCCTGTTCTCATTGACATCATTCGTAGAATCGAAGATAATCCTGATGTGAACATGGACTTAAGACAACTGTCCGCGTTACTTCTTCACACGTACCGCCAAGATGGTATAGAATTTCACCAGCCGGAAGGTAATTTAGCCATATCTTCGAATGTGATACCGTTTGCACCAACCTTCCACTCCTTTCATCGTCACAGGCTGCTACTAACAAGACTCATACCTAGTAATTTGCAGACACTTGGAAATACAACTGTTAATTCAGTGCTCAAGTGCACTCTACATCACATGTTGTCGACAACCGTTGACGCAAGACTGAGAGGAGATGAAAGCAATTGTAACCAGTTGTCACAGTATCGTGCGCTAAGAACAGGACGATCAATTAGCGATGACGTTGAAATTTTAGATACTTCCTTACTAAAAACTAATGATAAAAATGGGCAAATGCGGAATTTTGATCCAAACAGTGACGTTGAGTATACTAGTAATTACGGTAGTCTTAAATCTGAACGCCAATTACTCGGAGAAAGTCAATGCCCACTATTAGATGGAGTGATTTTCACCCAATGGGGAGCTGTATCAGGGGGAAACCTCATAGCTGGTATTGCGGCAGGTGCTCAACCACAACAGGTTCCTGTTCTTGAACTTGCAAAAGGATCTGTTTTAAATTATCCGAATGTCCAACAAACTGTGACTTCACTGTATCCAGCAACACTCTCAGGTGATTTGGCCGAGGCAATTCTTATTCAAGGAACTGAAAGAGGCAGTTCATCAATTTCGATCGGTACAGCAGGAAACTGGAACAGCACTCAAGCGCCTAGACACTTCATGATTCACAGCCGTGTCAACATAGAAATGACCGATCCAGAAATCAGAGGAGGCATCGATGGCTTTGTTCTCGGCAGTGCAATAACCTCAGTTTTAGGAACTTTTAGTTCTTTGAGACTGTCGCAGCTGTTGGACATGTATTATACACCGCGGAATGGGATATTTAGTCAAGCTTTTCGTGCATGTAACCGTCGCGAATTGAGTCAAACGCATATCACAAATCAAAATTTAGCTGGGGAAACTTTTGCCTTTGCTGCCGCTCTTGATACGAACATGCCACTTAGAGGAACTATAATGGGTGGCCTCGAGCAACTAGTGAACAGCGCAGTCACCAATTTCCAAACATATTCTACTAGCAATTTGAACGACTTAAGCTGTGTTACCTCAGTAGCAACGAGCCTGGATTACAGATTAAAAACGAATTTGTATATAGTTCTTGACTCATCATGGCCATACCAAGCAGTGTATCCGGCCATTGCATATTTAGTGGATTCAATAGAAGTAGACAAATTCGGATCGAGCATCACCCTGCTCAATGCATTTGATGGTAGTGTTGTTGTCAACACAACTTTCTCGCAAGTTGAATTCCACTCTAATTACACTTTAGCGGCACATCAATCAATGCTCAATGGAGTAAATCTGCAAACTAGTCTGACAAACATCAGAATAATGATGCAAGACCAACTTCGTAATGAAAGCGCAAGAAATTACGTAGGAGGAAATTCAACTGTAGTGTTGTACCTCATCAATACCGGTCATCTACAGAACAATGAAATAGTTTTTGAACAAGCCCGACTTCTTAATGAAACAGTTCCGGATTTGAGGCTACTTTTTGCGACTGCTACGAATCAGTTCGATAACATATGGAATTTGGTGCGTGATATTCACAACGACATCAGGGCGGTATCTTTGGCTACAACAGGCACAAACGTACCAATAGTCATGAGCCCAGTCCTGGAAAGAATTCAGTCAGTTGGAAGAAGAATTGTGAATCCTCTTTGTGGTTCACAGTTTGTTGATCAATTGTCAGGCACGAGACAGTTTGACGACTTCGTGGAACCTGGTTATATTAATTTCTACACAGTTAGTCCCAATTACTTTTATGTACACAATGATAACAGAAGAATCAGAATTTCTCGAAGTGCGAGTGGGGCTGGAAGTCTTGTTGTGTGTCATTCAAGAGCTGTTGCTCAACCAAGACGCAACACAACAATAGTAGGACTTGATCCAAGCGCAATAACTTGTGAGAATTTGGCTTCCACCGGAAATATTGAAATAAGGCTGCAGAATGCTTGCGACAGTTTTTGGACCATAGGATCATGTCCACCACTACACATTTCCGTACAATCAGAAACTGGTGGTGTTGATGCCACAAGCGCGATTTGCACAGATGCTTCGGTATGCAGATTCCCATACAACATTAGATATCAAGTCCAAATTGAAGACCTGGGGTGTTTTAGTGGAGCATCAAGTCTAGGAGCCAGCCTCGCAATTATTCTGACTTCTTTGTATCTCGCAATATTCTAA
Protein
MYWLPFFMVAVVGTTHGQLTAQSSIAPELRECYFDPILLNRNNLPPTTIPVLIDIIRRIEDNPDVNMDLRQLSALLLHTYRQDGIEFHQPEGNLAISSNVIPFAPTFHSFHRHRLLLTRLIPSNLQTLGNTTVNSVLKCTLHHMLSTTVDARLRGDESNCNQLSQYRALRTGRSISDDVEILDTSLLKTNDKNGQMRNFDPNSDVEYTSNYGSLKSERQLLGESQCPLLDGVIFTQWGAVSGGNLIAGIAAGAQPQQVPVLELAKGSVLNYPNVQQTVTSLYPATLSGDLAEAILIQGTERGSSSISIGTAGNWNSTQAPRHFMIHSRVNIEMTDPEIRGGIDGFVLGSAITSVLGTFSSLRLSQLLDMYYTPRNGIFSQAFRACNRRELSQTHITNQNLAGETFAFAAALDTNMPLRGTIMGGLEQLVNSAVTNFQTYSTSNLNDLSCVTSVATSLDYRLKTNLYIVLDSSWPYQAVYPAIAYLVDSIEVDKFGSSITLLNAFDGSVVVNTTFSQVEFHSNYTLAAHQSMLNGVNLQTSLTNIRIMMQDQLRNESARNYVGGNSTVVLYLINTGHLQNNEIVFEQARLLNETVPDLRLLFATATNQFDNIWNLVRDIHNDIRAVSLATTGTNVPIVMSPVLERIQSVGRRIVNPLCGSQFVDQLSGTRQFDDFVEPGYINFYTVSPNYFYVHNDNRRIRISRSASGAGSLVVCHSRAVAQPRRNTTIVGLDPSAITCENLASTGNIEIRLQNACDSFWTIGSCPPLHISVQSETGGVDATSAICTDASVCRFPYNIRYQVQIEDLGCFSGASSLGASLAIILTSLYLAIF
Summary
Uniprot
H9J1S1
A0A194RHA3
A0A194PNC2
A0A2A4K3Q3
A0A2A4K302
A0A2A4K2A9
+ More
A0A0L7L584 A0A212FBC0 A0A2H1W007 A0A1S4FTI5 Q16PR4 A0A1L8D9X5 A0A1L8D9M0 A0A182YPE2 A0A084WBC6 A0A182WF71 A0A182HNJ0 A0A182XC88 Q7Q343 A0A1Q3FIV2 A0A1Q3FJ36 A0A1Q3FKZ8 A0A1Q3FKQ3 A0A1Q3FK80 A0A1Q3FK23 B0W770 A0A1L8EC68 A0A1L8ECG4 A0A1A9W7T5 B4KSS8 Q28Z84 A0A0K8W4B4 A0A0A1XL35 A0A336MP36 A0A3B0IY76 A0A0M3QVR1 A0A1I8NTG3 A0A1I8N3I3 A0A336MV53 B4GI45 A0A034VDF0 B4PC03 U5ESY0 A0A0R1DZL7 A0A1A9VVM6 B4LNT2 A0A1B0G4K1 A0A0Q9W2F4 A0A0J9RKN8 B4QD68 A0A1A9Z5D9 Q8MLN4 Q6NP37 B3NRB0 W8BJC0 B4MPL3 B3MCE5 B4JVJ1 A0A1A9XHF3 A0A1Y1NED5 A0A1J1ISM2 A0A0T6AUX0 A0A2A3EMU3 A0A1W4V5T1 A0A0M8ZWE0 A0A154PRG2 A0A182LPH2 A0A088A0Z6 A0A1I8NTG2 A0A232EZ46 B4IHD6 A0A0J9RM94 A0A0R1DU80 A0A0R1DXJ9 Q0E8V8 A0A182VJ84 A0A0L7QUH1 A0A182QR20 A0A182TU21 A0A182R8N9 A0A182J8Y4 A0A182FPP0 A0A1B6F181 W5JAK7 A0A182JV63 A0A182MR66 A0A0R1E0C9 A0A1B6D0F0 A0A182P9B3 A0A158P2R3 A0A026VV20 V5GXA9 D6W7I5 A0A195DCI5 A0A195BBY9 A0A182MXZ9 F4WM87 E2A0U6
A0A0L7L584 A0A212FBC0 A0A2H1W007 A0A1S4FTI5 Q16PR4 A0A1L8D9X5 A0A1L8D9M0 A0A182YPE2 A0A084WBC6 A0A182WF71 A0A182HNJ0 A0A182XC88 Q7Q343 A0A1Q3FIV2 A0A1Q3FJ36 A0A1Q3FKZ8 A0A1Q3FKQ3 A0A1Q3FK80 A0A1Q3FK23 B0W770 A0A1L8EC68 A0A1L8ECG4 A0A1A9W7T5 B4KSS8 Q28Z84 A0A0K8W4B4 A0A0A1XL35 A0A336MP36 A0A3B0IY76 A0A0M3QVR1 A0A1I8NTG3 A0A1I8N3I3 A0A336MV53 B4GI45 A0A034VDF0 B4PC03 U5ESY0 A0A0R1DZL7 A0A1A9VVM6 B4LNT2 A0A1B0G4K1 A0A0Q9W2F4 A0A0J9RKN8 B4QD68 A0A1A9Z5D9 Q8MLN4 Q6NP37 B3NRB0 W8BJC0 B4MPL3 B3MCE5 B4JVJ1 A0A1A9XHF3 A0A1Y1NED5 A0A1J1ISM2 A0A0T6AUX0 A0A2A3EMU3 A0A1W4V5T1 A0A0M8ZWE0 A0A154PRG2 A0A182LPH2 A0A088A0Z6 A0A1I8NTG2 A0A232EZ46 B4IHD6 A0A0J9RM94 A0A0R1DU80 A0A0R1DXJ9 Q0E8V8 A0A182VJ84 A0A0L7QUH1 A0A182QR20 A0A182TU21 A0A182R8N9 A0A182J8Y4 A0A182FPP0 A0A1B6F181 W5JAK7 A0A182JV63 A0A182MR66 A0A0R1E0C9 A0A1B6D0F0 A0A182P9B3 A0A158P2R3 A0A026VV20 V5GXA9 D6W7I5 A0A195DCI5 A0A195BBY9 A0A182MXZ9 F4WM87 E2A0U6
Pubmed
19121390
26354079
26227816
22118469
17510324
25244985
+ More
24438588 12364791 14747013 17210077 17994087 15632085 25830018 25315136 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 20966253 28648823 20920257 23761445 21347285 24508170 30249741 18362917 19820115 21719571 20798317
24438588 12364791 14747013 17210077 17994087 15632085 25830018 25315136 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 20966253 28648823 20920257 23761445 21347285 24508170 30249741 18362917 19820115 21719571 20798317
EMBL
BABH01008108
KQ460207
KPJ16700.1
KQ459597
KPI94817.1
NWSH01000208
+ More
PCG78393.1 PCG78396.1 PCG78395.1 JTDY01002864 KOB70580.1 AGBW02009362 OWR51017.1 ODYU01005427 SOQ46256.1 CH477774 EAT36358.1 GFDF01010919 JAV03165.1 GFDF01010918 JAV03166.1 ATLV01022343 KE525331 KFB47520.1 APCN01001936 AAAB01008966 EAA13062.4 GFDL01007591 JAV27454.1 GFDL01007510 JAV27535.1 GFDL01006871 JAV28174.1 GFDL01006888 JAV28157.1 GFDL01007077 JAV27968.1 GFDL01007202 JAV27843.1 DS231852 EDS37675.1 GFDG01002462 JAV16337.1 GFDG01002463 JAV16336.1 CH933808 EDW10577.2 CM000071 EAL25730.4 GDHF01006717 GDHF01006604 JAI45597.1 JAI45710.1 GBXI01003044 JAD11248.1 UFQT01001909 SSX32132.1 OUUW01000001 SPP72657.1 CP012524 ALC42833.1 UFQT01003040 SSX34484.1 CH479183 EDW36165.1 GAKP01018418 JAC40534.1 CM000158 EDW92657.1 KRK00630.1 GANO01004630 JAB55241.1 KRK00628.1 CH940648 EDW60152.2 CCAG010000826 KRF79283.1 CM002911 KMY96533.1 CM000362 EDX08664.1 AE013599 AAM68331.2 AGB93709.1 AGB93710.1 BT011094 AAR82760.1 CH954179 EDV57129.1 GAMC01016857 GAMC01016856 JAB89699.1 CH963849 EDW74052.1 CH902619 EDV37269.1 CH916375 EDV98459.1 GEZM01007311 JAV95250.1 CVRI01000059 CRL03128.1 LJIG01022756 KRT78890.1 KZ288215 PBC32582.1 KQ435821 KOX72475.1 KQ435078 KZC14463.1 NNAY01001572 OXU23557.1 CH480838 EDW49312.1 KMY96534.1 KRK00629.1 KRK00627.1 BT083392 ABI31116.2 ACQ66053.1 KQ414735 KOC62307.1 AXCN02001648 GECZ01025851 JAS43918.1 ADMH02001669 ETN61482.1 AXCM01002130 KRK00631.1 GEDC01030956 GEDC01018116 JAS06342.1 JAS19182.1 ADTU01001291 KK107796 QOIP01000013 EZA47608.1 RLU15414.1 GALX01003548 JAB64918.1 KQ971307 EFA11281.1 KQ980989 KYN10547.1 KQ976522 KYM82068.1 GL888217 EGI64731.1 GL435626 EFN72999.1
PCG78393.1 PCG78396.1 PCG78395.1 JTDY01002864 KOB70580.1 AGBW02009362 OWR51017.1 ODYU01005427 SOQ46256.1 CH477774 EAT36358.1 GFDF01010919 JAV03165.1 GFDF01010918 JAV03166.1 ATLV01022343 KE525331 KFB47520.1 APCN01001936 AAAB01008966 EAA13062.4 GFDL01007591 JAV27454.1 GFDL01007510 JAV27535.1 GFDL01006871 JAV28174.1 GFDL01006888 JAV28157.1 GFDL01007077 JAV27968.1 GFDL01007202 JAV27843.1 DS231852 EDS37675.1 GFDG01002462 JAV16337.1 GFDG01002463 JAV16336.1 CH933808 EDW10577.2 CM000071 EAL25730.4 GDHF01006717 GDHF01006604 JAI45597.1 JAI45710.1 GBXI01003044 JAD11248.1 UFQT01001909 SSX32132.1 OUUW01000001 SPP72657.1 CP012524 ALC42833.1 UFQT01003040 SSX34484.1 CH479183 EDW36165.1 GAKP01018418 JAC40534.1 CM000158 EDW92657.1 KRK00630.1 GANO01004630 JAB55241.1 KRK00628.1 CH940648 EDW60152.2 CCAG010000826 KRF79283.1 CM002911 KMY96533.1 CM000362 EDX08664.1 AE013599 AAM68331.2 AGB93709.1 AGB93710.1 BT011094 AAR82760.1 CH954179 EDV57129.1 GAMC01016857 GAMC01016856 JAB89699.1 CH963849 EDW74052.1 CH902619 EDV37269.1 CH916375 EDV98459.1 GEZM01007311 JAV95250.1 CVRI01000059 CRL03128.1 LJIG01022756 KRT78890.1 KZ288215 PBC32582.1 KQ435821 KOX72475.1 KQ435078 KZC14463.1 NNAY01001572 OXU23557.1 CH480838 EDW49312.1 KMY96534.1 KRK00629.1 KRK00627.1 BT083392 ABI31116.2 ACQ66053.1 KQ414735 KOC62307.1 AXCN02001648 GECZ01025851 JAS43918.1 ADMH02001669 ETN61482.1 AXCM01002130 KRK00631.1 GEDC01030956 GEDC01018116 JAS06342.1 JAS19182.1 ADTU01001291 KK107796 QOIP01000013 EZA47608.1 RLU15414.1 GALX01003548 JAB64918.1 KQ971307 EFA11281.1 KQ980989 KYN10547.1 KQ976522 KYM82068.1 GL888217 EGI64731.1 GL435626 EFN72999.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000008820 UP000076408 UP000030765 UP000075920 UP000075840 UP000076407 UP000007062 UP000002320 UP000091820 UP000009192 UP000001819 UP000268350 UP000092553 UP000095300 UP000095301 UP000008744 UP000002282 UP000078200 UP000008792 UP000092444 UP000000304 UP000092445 UP000000803 UP000008711 UP000007798 UP000007801 UP000001070 UP000092443 UP000183832 UP000242457 UP000192221 UP000053105 UP000076502 UP000075882 UP000005203 UP000215335 UP000001292 UP000075903 UP000053825 UP000075886 UP000075902 UP000075900 UP000075880 UP000069272 UP000000673 UP000075881 UP000075883 UP000075885 UP000005205 UP000053097 UP000279307 UP000007266 UP000078492 UP000078540 UP000075884 UP000007755 UP000000311
UP000008820 UP000076408 UP000030765 UP000075920 UP000075840 UP000076407 UP000007062 UP000002320 UP000091820 UP000009192 UP000001819 UP000268350 UP000092553 UP000095300 UP000095301 UP000008744 UP000002282 UP000078200 UP000008792 UP000092444 UP000000304 UP000092445 UP000000803 UP000008711 UP000007798 UP000007801 UP000001070 UP000092443 UP000183832 UP000242457 UP000192221 UP000053105 UP000076502 UP000075882 UP000005203 UP000215335 UP000001292 UP000075903 UP000053825 UP000075886 UP000075902 UP000075900 UP000075880 UP000069272 UP000000673 UP000075881 UP000075883 UP000075885 UP000005205 UP000053097 UP000279307 UP000007266 UP000078492 UP000078540 UP000075884 UP000007755 UP000000311
ProteinModelPortal
H9J1S1
A0A194RHA3
A0A194PNC2
A0A2A4K3Q3
A0A2A4K302
A0A2A4K2A9
+ More
A0A0L7L584 A0A212FBC0 A0A2H1W007 A0A1S4FTI5 Q16PR4 A0A1L8D9X5 A0A1L8D9M0 A0A182YPE2 A0A084WBC6 A0A182WF71 A0A182HNJ0 A0A182XC88 Q7Q343 A0A1Q3FIV2 A0A1Q3FJ36 A0A1Q3FKZ8 A0A1Q3FKQ3 A0A1Q3FK80 A0A1Q3FK23 B0W770 A0A1L8EC68 A0A1L8ECG4 A0A1A9W7T5 B4KSS8 Q28Z84 A0A0K8W4B4 A0A0A1XL35 A0A336MP36 A0A3B0IY76 A0A0M3QVR1 A0A1I8NTG3 A0A1I8N3I3 A0A336MV53 B4GI45 A0A034VDF0 B4PC03 U5ESY0 A0A0R1DZL7 A0A1A9VVM6 B4LNT2 A0A1B0G4K1 A0A0Q9W2F4 A0A0J9RKN8 B4QD68 A0A1A9Z5D9 Q8MLN4 Q6NP37 B3NRB0 W8BJC0 B4MPL3 B3MCE5 B4JVJ1 A0A1A9XHF3 A0A1Y1NED5 A0A1J1ISM2 A0A0T6AUX0 A0A2A3EMU3 A0A1W4V5T1 A0A0M8ZWE0 A0A154PRG2 A0A182LPH2 A0A088A0Z6 A0A1I8NTG2 A0A232EZ46 B4IHD6 A0A0J9RM94 A0A0R1DU80 A0A0R1DXJ9 Q0E8V8 A0A182VJ84 A0A0L7QUH1 A0A182QR20 A0A182TU21 A0A182R8N9 A0A182J8Y4 A0A182FPP0 A0A1B6F181 W5JAK7 A0A182JV63 A0A182MR66 A0A0R1E0C9 A0A1B6D0F0 A0A182P9B3 A0A158P2R3 A0A026VV20 V5GXA9 D6W7I5 A0A195DCI5 A0A195BBY9 A0A182MXZ9 F4WM87 E2A0U6
A0A0L7L584 A0A212FBC0 A0A2H1W007 A0A1S4FTI5 Q16PR4 A0A1L8D9X5 A0A1L8D9M0 A0A182YPE2 A0A084WBC6 A0A182WF71 A0A182HNJ0 A0A182XC88 Q7Q343 A0A1Q3FIV2 A0A1Q3FJ36 A0A1Q3FKZ8 A0A1Q3FKQ3 A0A1Q3FK80 A0A1Q3FK23 B0W770 A0A1L8EC68 A0A1L8ECG4 A0A1A9W7T5 B4KSS8 Q28Z84 A0A0K8W4B4 A0A0A1XL35 A0A336MP36 A0A3B0IY76 A0A0M3QVR1 A0A1I8NTG3 A0A1I8N3I3 A0A336MV53 B4GI45 A0A034VDF0 B4PC03 U5ESY0 A0A0R1DZL7 A0A1A9VVM6 B4LNT2 A0A1B0G4K1 A0A0Q9W2F4 A0A0J9RKN8 B4QD68 A0A1A9Z5D9 Q8MLN4 Q6NP37 B3NRB0 W8BJC0 B4MPL3 B3MCE5 B4JVJ1 A0A1A9XHF3 A0A1Y1NED5 A0A1J1ISM2 A0A0T6AUX0 A0A2A3EMU3 A0A1W4V5T1 A0A0M8ZWE0 A0A154PRG2 A0A182LPH2 A0A088A0Z6 A0A1I8NTG2 A0A232EZ46 B4IHD6 A0A0J9RM94 A0A0R1DU80 A0A0R1DXJ9 Q0E8V8 A0A182VJ84 A0A0L7QUH1 A0A182QR20 A0A182TU21 A0A182R8N9 A0A182J8Y4 A0A182FPP0 A0A1B6F181 W5JAK7 A0A182JV63 A0A182MR66 A0A0R1E0C9 A0A1B6D0F0 A0A182P9B3 A0A158P2R3 A0A026VV20 V5GXA9 D6W7I5 A0A195DCI5 A0A195BBY9 A0A182MXZ9 F4WM87 E2A0U6
Ontologies
GO
PANTHER
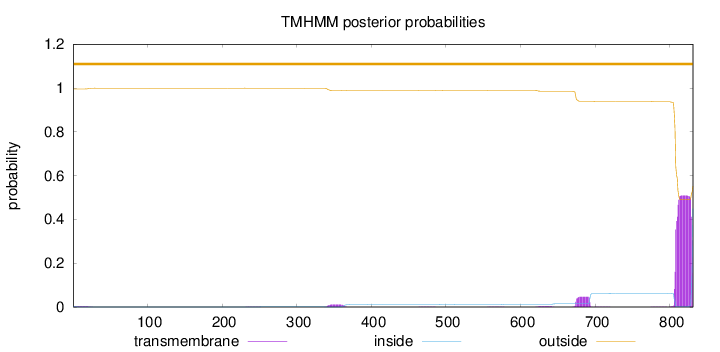
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.993167
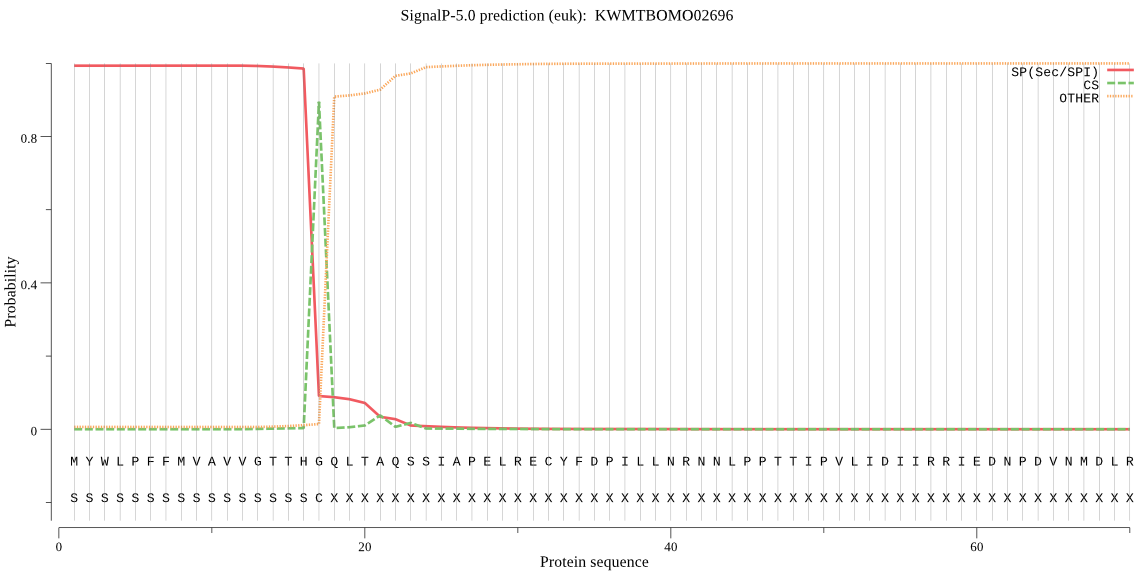
Length:
831
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.49838
Exp number, first 60 AAs:
0.06889
Total prob of N-in:
0.00405
outside
1 - 831
Population Genetic Test Statistics
Pi
274.518875
Theta
192.845115
Tajima's D
1.468728
CLR
0.175226
CSRT
0.779511024448778
Interpretation
Uncertain
