Pre Gene Modal
BGIBMGA003457
Annotation
ras-related_protein_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.548
Sequence
CDS
ATGCGCGAATTTAAAGTGGTCGTGTTGGGTTCGGGTGGGGTCGGAAAGAGTGCGTTGACTGTGCAATTTGTATCAGGATGTTTTATGGAAAAATATGATCCGACGATAGAGGATTTCTACAGAAAAGAAATCGAAGTGGACAATTCTCCTTGTGTTTTAGAAATTTTGGACACCGCTGGCACGGAGCAGTTTGCTTCGATGCGAGATCTGTATATAAAAAATGGTCAAGGCTTTGTAGTAGTATATTCATTAACAAACCACCAAACGTTTCAAGACATCAAACCAATGAAGGAATTGATTACTCGTGTCAAAGGATCAGAACGTGTACCAATTCTACTTGTGGGAAATAAGGCTGACTTGGAACATCAACGTGAGGTGGCGCATGCGGAGGGCGCTGCGCTCGCGCAGATGTGGGGCTGTCCCTTCGTCGAAGCTTCAGCAAAGAGTCGCACCAACGTGAACGAGATGTTTGCAGAAATTGTGCGTGAAATGAACGTTAGTCCAGAAAAAGAAAAACCTTATTGTTGTTGTACAGTGCTTTAA
Protein
MREFKVVVLGSGGVGKSALTVQFVSGCFMEKYDPTIEDFYRKEIEVDNSPCVLEILDTAGTEQFASMRDLYIKNGQGFVVVYSLTNHQTFQDIKPMKELITRVKGSERVPILLVGNKADLEHQREVAHAEGAALAQMWGCPFVEASAKSRTNVNEMFAEIVREMNVSPEKEKPYCCCTVL
Summary
Uniprot
Q2F5M0
A0A2A4JD26
A0A2H1W197
S4Q065
A0A212FBB3
A0A0L7LM15
+ More
A0A1E1VZM3 A0A1B6C8R8 A0A1B6JUT3 A0A1B6MHK9 A0A0A9YZG0 A0A0L7QTY3 A0A232F838 A0A0P4VST0 A0A023F5V6 T1IB37 A0A2P8YA32 A0A0C9Q348 A0A2R7X6D1 A0A2A3EFH6 V9IL12 A0A0M9ABI6 A0A088ARZ9 A0A1W4WR34 A0A154PGV4 A0A026VU48 A0A195EJH3 E9ILH1 A0A195BFU9 A0A158NJM6 A0A286NL57 E2ATE5 A0A151X6J2 A0A195FNA2 A0A151ICC4 J3JZC0 D6W8A5 A0A2S2QMH2 A0A1Y1MUP3 A0A2H8TSZ3 J9K922 A0A2S2NQF0 A0A194RL39 A0A194PQ01 A0A067QGM5 A0A0P4WSW3 E9GIA3 A0A164K3F9 E2BYE9 A0A2P2I6P6 A0A0P6AHH7 A0A0J7L6E1 K7IUI3 A0A1S3DCU7 T1IN97 A0A224Y0H5 A0A1J1HIG2 A0A2P6L6W4 A0A087UPX2 A0A1Q3EUC4 B4LP06 A0A1Y3B5A2 A0A1A9XZM3 A0A1A9V6H8 A0A1B0B9Z2 A0A1A9W117 D3TQW7 B4J673 T1E329 T1E937 A0A2M4CSV2 A0A2M3ZAT0 A0A2M4ATJ9 A0A131ZXK2 A0A1W4V0H4 B5E0W7 B4GCJ4 B4PAT3 B3MDX3 B4I2C5 B3NQ70 B4QBI6 O96692 A0A023EIF7 U5EVB4 A0A0K8UG43 A0A034WW21 A0A0A1WMP3 B4KLX6 A0A1I8PRB7 A0A084VZR3 Q7PMT0 A0A1L8DV40 A0A023GFH4 G3MNM5 A0A1E1XU85 A0A023FG93 A0A023FU98 A0A1E1X0S1 A0A1J1HJW8
A0A1E1VZM3 A0A1B6C8R8 A0A1B6JUT3 A0A1B6MHK9 A0A0A9YZG0 A0A0L7QTY3 A0A232F838 A0A0P4VST0 A0A023F5V6 T1IB37 A0A2P8YA32 A0A0C9Q348 A0A2R7X6D1 A0A2A3EFH6 V9IL12 A0A0M9ABI6 A0A088ARZ9 A0A1W4WR34 A0A154PGV4 A0A026VU48 A0A195EJH3 E9ILH1 A0A195BFU9 A0A158NJM6 A0A286NL57 E2ATE5 A0A151X6J2 A0A195FNA2 A0A151ICC4 J3JZC0 D6W8A5 A0A2S2QMH2 A0A1Y1MUP3 A0A2H8TSZ3 J9K922 A0A2S2NQF0 A0A194RL39 A0A194PQ01 A0A067QGM5 A0A0P4WSW3 E9GIA3 A0A164K3F9 E2BYE9 A0A2P2I6P6 A0A0P6AHH7 A0A0J7L6E1 K7IUI3 A0A1S3DCU7 T1IN97 A0A224Y0H5 A0A1J1HIG2 A0A2P6L6W4 A0A087UPX2 A0A1Q3EUC4 B4LP06 A0A1Y3B5A2 A0A1A9XZM3 A0A1A9V6H8 A0A1B0B9Z2 A0A1A9W117 D3TQW7 B4J673 T1E329 T1E937 A0A2M4CSV2 A0A2M3ZAT0 A0A2M4ATJ9 A0A131ZXK2 A0A1W4V0H4 B5E0W7 B4GCJ4 B4PAT3 B3MDX3 B4I2C5 B3NQ70 B4QBI6 O96692 A0A023EIF7 U5EVB4 A0A0K8UG43 A0A034WW21 A0A0A1WMP3 B4KLX6 A0A1I8PRB7 A0A084VZR3 Q7PMT0 A0A1L8DV40 A0A023GFH4 G3MNM5 A0A1E1XU85 A0A023FG93 A0A023FU98 A0A1E1X0S1 A0A1J1HJW8
Pubmed
19121390
23622113
22118469
26227816
25401762
26823975
+ More
28648823 27129103 25474469 29403074 24508170 30249741 21282665 21347285 20798317 22516182 23537049 18362917 19820115 28004739 26354079 24845553 21292972 20075255 17994087 20353571 24330624 26555130 15632085 17550304 22936249 9927464 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 25348373 25830018 24438588 12364791 14747013 17210077 22216098 29209593 28503490
28648823 27129103 25474469 29403074 24508170 30249741 21282665 21347285 20798317 22516182 23537049 18362917 19820115 28004739 26354079 24845553 21292972 20075255 17994087 20353571 24330624 26555130 15632085 17550304 22936249 9927464 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 25348373 25830018 24438588 12364791 14747013 17210077 22216098 29209593 28503490
EMBL
BABH01008121
DQ311403
ABD36347.1
NWSH01002038
PCG69340.1
ODYU01005650
+ More
SOQ46716.1 GAIX01000328 JAA92232.1 AGBW02009362 OWR51020.1 JTDY01000579 KOB76593.1 GDQN01010868 JAT80186.1 GEDC01027579 GEDC01023611 GEDC01007988 GEDC01001011 JAS09719.1 JAS13687.1 JAS29310.1 JAS36287.1 GECU01004770 JAT02937.1 GEBQ01021977 GEBQ01004539 JAT18000.1 JAT35438.1 GBHO01005192 GBRD01008461 GDHC01016174 JAG38412.1 JAG57360.1 JAQ02455.1 KQ414736 KOC62122.1 NNAY01000680 OXU27016.1 GDKW01000945 JAI55650.1 GBBI01002144 JAC16568.1 ACPB03021689 PYGN01000763 PSN41101.1 GBYB01008503 JAG78270.1 KK857617 PTY27347.1 KZ288271 PBC29741.1 JR052602 AEY61735.1 KQ435710 KOX79883.1 KQ434892 KZC10548.1 KK107928 QOIP01000001 EZA47165.1 RLU27053.1 RLU27248.1 KQ978782 KYN28400.1 GL764076 EFZ18586.1 KQ976490 KYM83460.1 ADTU01018071 KX809579 ASZ69800.1 GL442548 EFN63312.1 KQ982482 KYQ55954.1 KQ981424 KYN41812.1 KQ978058 KYM97743.1 APGK01020127 BT128601 KB740167 KB631669 AEE63558.1 ENN81199.1 ERL85166.1 KQ971307 EFA10889.1 GGMS01009752 MBY78955.1 GEZM01021648 JAV88888.1 GFXV01005166 MBW16971.1 ABLF02021004 GGMR01006719 MBY19338.1 KQ460207 KPJ16696.1 KQ459597 KPI94814.1 KK853500 KDR07159.1 GDRN01064086 JAI64891.1 GL732546 EFX80641.1 LRGB01003375 KZS02913.1 GL451460 EFN79281.1 IACF01003947 LAB69550.1 GDIP01029213 JAM74502.1 LBMM01000532 KMQ98123.1 AAZX01002638 JH431152 GFTR01002515 JAW13911.1 CVRI01000006 CRK87840.1 MWRG01001367 PRD34333.1 KK120932 KFM79411.1 GFDL01016189 JAV18856.1 CH940648 EDW61175.1 MUJZ01039392 OTF76022.1 JXJN01010648 JXJN01010649 CCAG010021563 EZ423819 ADD20095.1 CH916367 EDW01931.1 GALA01000745 JAA94107.1 GAMD01002203 JAA99387.1 GGFL01004225 MBW68403.1 GGFM01004888 MBW25639.1 GGFK01010711 MBW44032.1 JXLN01004942 KPM03413.1 CM000071 EDY69048.1 CH479181 EDW32471.1 CM000158 EDW92473.1 CH902619 EDV37518.1 CH480820 EDW53920.1 CH954179 EDV56943.1 CM000362 CM002911 EDX08504.1 KMY96245.1 AF111427 AE013599 BT012453 AAC98693.1 AAF47155.1 AAS93724.1 GAPW01004878 GEHC01000772 JAC08720.1 JAV46873.1 GANO01001952 JAB57919.1 GDHF01026667 JAI25647.1 GAKP01000148 JAC58804.1 GBXI01014527 JAC99764.1 CH933808 EDW09786.1 ATLV01018944 ATLV01018945 ATLV01018946 KE525256 KFB43457.1 AAAB01008968 EAA13245.4 GFDF01003794 JAV10290.1 GBBM01002761 JAC32657.1 JO843476 AEO35093.1 GFAA01000574 JAU02861.1 GBBK01003940 JAC20542.1 GBBL01002068 JAC25252.1 GFAC01006315 JAT92873.1 CRK87842.1 CRK87843.1
SOQ46716.1 GAIX01000328 JAA92232.1 AGBW02009362 OWR51020.1 JTDY01000579 KOB76593.1 GDQN01010868 JAT80186.1 GEDC01027579 GEDC01023611 GEDC01007988 GEDC01001011 JAS09719.1 JAS13687.1 JAS29310.1 JAS36287.1 GECU01004770 JAT02937.1 GEBQ01021977 GEBQ01004539 JAT18000.1 JAT35438.1 GBHO01005192 GBRD01008461 GDHC01016174 JAG38412.1 JAG57360.1 JAQ02455.1 KQ414736 KOC62122.1 NNAY01000680 OXU27016.1 GDKW01000945 JAI55650.1 GBBI01002144 JAC16568.1 ACPB03021689 PYGN01000763 PSN41101.1 GBYB01008503 JAG78270.1 KK857617 PTY27347.1 KZ288271 PBC29741.1 JR052602 AEY61735.1 KQ435710 KOX79883.1 KQ434892 KZC10548.1 KK107928 QOIP01000001 EZA47165.1 RLU27053.1 RLU27248.1 KQ978782 KYN28400.1 GL764076 EFZ18586.1 KQ976490 KYM83460.1 ADTU01018071 KX809579 ASZ69800.1 GL442548 EFN63312.1 KQ982482 KYQ55954.1 KQ981424 KYN41812.1 KQ978058 KYM97743.1 APGK01020127 BT128601 KB740167 KB631669 AEE63558.1 ENN81199.1 ERL85166.1 KQ971307 EFA10889.1 GGMS01009752 MBY78955.1 GEZM01021648 JAV88888.1 GFXV01005166 MBW16971.1 ABLF02021004 GGMR01006719 MBY19338.1 KQ460207 KPJ16696.1 KQ459597 KPI94814.1 KK853500 KDR07159.1 GDRN01064086 JAI64891.1 GL732546 EFX80641.1 LRGB01003375 KZS02913.1 GL451460 EFN79281.1 IACF01003947 LAB69550.1 GDIP01029213 JAM74502.1 LBMM01000532 KMQ98123.1 AAZX01002638 JH431152 GFTR01002515 JAW13911.1 CVRI01000006 CRK87840.1 MWRG01001367 PRD34333.1 KK120932 KFM79411.1 GFDL01016189 JAV18856.1 CH940648 EDW61175.1 MUJZ01039392 OTF76022.1 JXJN01010648 JXJN01010649 CCAG010021563 EZ423819 ADD20095.1 CH916367 EDW01931.1 GALA01000745 JAA94107.1 GAMD01002203 JAA99387.1 GGFL01004225 MBW68403.1 GGFM01004888 MBW25639.1 GGFK01010711 MBW44032.1 JXLN01004942 KPM03413.1 CM000071 EDY69048.1 CH479181 EDW32471.1 CM000158 EDW92473.1 CH902619 EDV37518.1 CH480820 EDW53920.1 CH954179 EDV56943.1 CM000362 CM002911 EDX08504.1 KMY96245.1 AF111427 AE013599 BT012453 AAC98693.1 AAF47155.1 AAS93724.1 GAPW01004878 GEHC01000772 JAC08720.1 JAV46873.1 GANO01001952 JAB57919.1 GDHF01026667 JAI25647.1 GAKP01000148 JAC58804.1 GBXI01014527 JAC99764.1 CH933808 EDW09786.1 ATLV01018944 ATLV01018945 ATLV01018946 KE525256 KFB43457.1 AAAB01008968 EAA13245.4 GFDF01003794 JAV10290.1 GBBM01002761 JAC32657.1 JO843476 AEO35093.1 GFAA01000574 JAU02861.1 GBBK01003940 JAC20542.1 GBBL01002068 JAC25252.1 GFAC01006315 JAT92873.1 CRK87842.1 CRK87843.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053825
UP000215335
+ More
UP000015103 UP000245037 UP000242457 UP000053105 UP000005203 UP000192223 UP000076502 UP000053097 UP000279307 UP000078492 UP000078540 UP000005205 UP000000311 UP000075809 UP000078541 UP000078542 UP000019118 UP000030742 UP000007266 UP000007819 UP000053240 UP000053268 UP000027135 UP000000305 UP000076858 UP000008237 UP000036403 UP000002358 UP000079169 UP000183832 UP000054359 UP000008792 UP000092443 UP000078200 UP000092460 UP000091820 UP000092444 UP000001070 UP000192221 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000095300 UP000030765 UP000007062
UP000015103 UP000245037 UP000242457 UP000053105 UP000005203 UP000192223 UP000076502 UP000053097 UP000279307 UP000078492 UP000078540 UP000005205 UP000000311 UP000075809 UP000078541 UP000078542 UP000019118 UP000030742 UP000007266 UP000007819 UP000053240 UP000053268 UP000027135 UP000000305 UP000076858 UP000008237 UP000036403 UP000002358 UP000079169 UP000183832 UP000054359 UP000008792 UP000092443 UP000078200 UP000092460 UP000091820 UP000092444 UP000001070 UP000192221 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000095300 UP000030765 UP000007062
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Q2F5M0
A0A2A4JD26
A0A2H1W197
S4Q065
A0A212FBB3
A0A0L7LM15
+ More
A0A1E1VZM3 A0A1B6C8R8 A0A1B6JUT3 A0A1B6MHK9 A0A0A9YZG0 A0A0L7QTY3 A0A232F838 A0A0P4VST0 A0A023F5V6 T1IB37 A0A2P8YA32 A0A0C9Q348 A0A2R7X6D1 A0A2A3EFH6 V9IL12 A0A0M9ABI6 A0A088ARZ9 A0A1W4WR34 A0A154PGV4 A0A026VU48 A0A195EJH3 E9ILH1 A0A195BFU9 A0A158NJM6 A0A286NL57 E2ATE5 A0A151X6J2 A0A195FNA2 A0A151ICC4 J3JZC0 D6W8A5 A0A2S2QMH2 A0A1Y1MUP3 A0A2H8TSZ3 J9K922 A0A2S2NQF0 A0A194RL39 A0A194PQ01 A0A067QGM5 A0A0P4WSW3 E9GIA3 A0A164K3F9 E2BYE9 A0A2P2I6P6 A0A0P6AHH7 A0A0J7L6E1 K7IUI3 A0A1S3DCU7 T1IN97 A0A224Y0H5 A0A1J1HIG2 A0A2P6L6W4 A0A087UPX2 A0A1Q3EUC4 B4LP06 A0A1Y3B5A2 A0A1A9XZM3 A0A1A9V6H8 A0A1B0B9Z2 A0A1A9W117 D3TQW7 B4J673 T1E329 T1E937 A0A2M4CSV2 A0A2M3ZAT0 A0A2M4ATJ9 A0A131ZXK2 A0A1W4V0H4 B5E0W7 B4GCJ4 B4PAT3 B3MDX3 B4I2C5 B3NQ70 B4QBI6 O96692 A0A023EIF7 U5EVB4 A0A0K8UG43 A0A034WW21 A0A0A1WMP3 B4KLX6 A0A1I8PRB7 A0A084VZR3 Q7PMT0 A0A1L8DV40 A0A023GFH4 G3MNM5 A0A1E1XU85 A0A023FG93 A0A023FU98 A0A1E1X0S1 A0A1J1HJW8
A0A1E1VZM3 A0A1B6C8R8 A0A1B6JUT3 A0A1B6MHK9 A0A0A9YZG0 A0A0L7QTY3 A0A232F838 A0A0P4VST0 A0A023F5V6 T1IB37 A0A2P8YA32 A0A0C9Q348 A0A2R7X6D1 A0A2A3EFH6 V9IL12 A0A0M9ABI6 A0A088ARZ9 A0A1W4WR34 A0A154PGV4 A0A026VU48 A0A195EJH3 E9ILH1 A0A195BFU9 A0A158NJM6 A0A286NL57 E2ATE5 A0A151X6J2 A0A195FNA2 A0A151ICC4 J3JZC0 D6W8A5 A0A2S2QMH2 A0A1Y1MUP3 A0A2H8TSZ3 J9K922 A0A2S2NQF0 A0A194RL39 A0A194PQ01 A0A067QGM5 A0A0P4WSW3 E9GIA3 A0A164K3F9 E2BYE9 A0A2P2I6P6 A0A0P6AHH7 A0A0J7L6E1 K7IUI3 A0A1S3DCU7 T1IN97 A0A224Y0H5 A0A1J1HIG2 A0A2P6L6W4 A0A087UPX2 A0A1Q3EUC4 B4LP06 A0A1Y3B5A2 A0A1A9XZM3 A0A1A9V6H8 A0A1B0B9Z2 A0A1A9W117 D3TQW7 B4J673 T1E329 T1E937 A0A2M4CSV2 A0A2M3ZAT0 A0A2M4ATJ9 A0A131ZXK2 A0A1W4V0H4 B5E0W7 B4GCJ4 B4PAT3 B3MDX3 B4I2C5 B3NQ70 B4QBI6 O96692 A0A023EIF7 U5EVB4 A0A0K8UG43 A0A034WW21 A0A0A1WMP3 B4KLX6 A0A1I8PRB7 A0A084VZR3 Q7PMT0 A0A1L8DV40 A0A023GFH4 G3MNM5 A0A1E1XU85 A0A023FG93 A0A023FU98 A0A1E1X0S1 A0A1J1HJW8
PDB
3RAP
E-value=4.85782e-60,
Score=581
Ontologies
GO
PANTHER
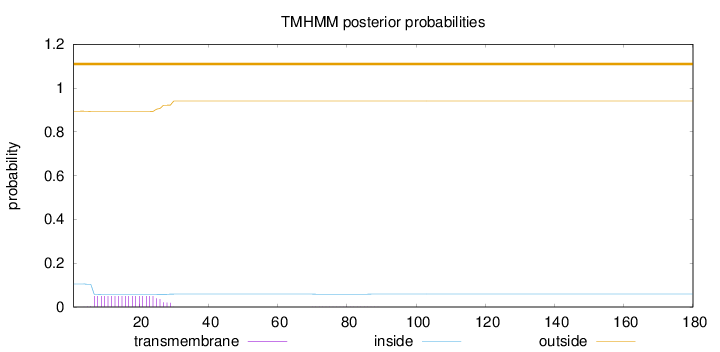
Topology
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.04615
Exp number, first 60 AAs:
1.03931
Total prob of N-in:
0.10555
outside
1 - 180
Population Genetic Test Statistics
Pi
313.163626
Theta
244.234097
Tajima's D
0.888991
CLR
0
CSRT
0.629468526573671
Interpretation
Uncertain
