Pre Gene Modal
BGIBMGA003744
Annotation
PREDICTED:_septin-7_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.316
Sequence
CDS
ATGAACGGCAGTACGACGACGGTAAATAGTGGTGCCGTCAACGGGCCTGGTGGTACCGTTAATGGAATCGGTGCTGCTCCACGTACAGCAACTCTTGGAACCCCAAGCAGCTATGCTTACACATCAAGCGGCTCATTACTGGGTCGAGGGGGCTCCGCTAGACCTGTACCACCTCCGACGCTACCAAAGTACAGTGGCTCTTTTAGTGCTGGAAGTACAACGGGTCTCCGAGAACGCGAAAGAGAAGGAACAGGTGGATCACATCGTTTGGCAAGCTTGGAAAGATTGGCTTTGCGCCAAAGGATTATTGAACAAAGTGCCAATAATCAGGTGGCGAGCGCAACAATTATCAATAGTTCTTCAAACGGAGGCCTCACTTCTGCTACAGTAACCCTCACCGATACTGCAACGTTACAGAGTAAACGAGAGCTGTTTTTCAAGTCGGACACTGTAAGCGGTGGCGGCGTAAGCGCGTCTGTCCCGCCTTCTGCAGCACCGCCCGCGCCTCCTTCCACGCTCAAAGATGCGCTCAGTAAGCGCTCTGCGACGCTACCAGAGCCTCAGGAGACTAACCACGTAGAAGCCAATGGGATCGCCACGAAACACAGCAATAGCGTGTCGGTAGATGTCTCCAAGCCTCCGAGTACAGCGCCCCCACCAACACCTGCCCCGCCAGTACCAATATCTGCACCTCCCGCACCAGTACCGACACCCCTCAGGAGTACAGAGAATATTATGAAGAAAGCCGAACACCCACCGGTCGCGCCGAAACCAGACTTACCTAAAATAGAAAAACCAAAAACCAAAGAACTAGATGGCTACGTCGGTTTTGCTAATCTGCCGAACCAAGTGTACAGGAAAGCTGTGAAGAAGGGCTTCGAATTCACGTTGATGGTCGTAGGTGAATCAGGCTTGGGCAAGTCGACACTAATAAACTCGCTCTTCTTGACGGAGGTGTATGACAAGGACAAACATCCGGGGCCGTCGCTGCGAGTGAAGAAGACGGTTGGCGTTGAAACCAGCGTCGTTCTTCTAAAAGAGAATGGTGTTAACCTCACGCTGACCATCGTCGACACGCCGGGATTCGGCGACGCCGTCGATAACAGTAATTGTTGGCAACCGATCATCGACTTCGTTGAATCGAAGTACGAAGAGTTTTTGAACGCCGAATCACGTGTCGCCCGCAAGGCGGCGCCACCGGACACGCGCGTGCACTGCTGCCTGTACTTCATAGCGCCGAGCGGGCACGGACTCAAACCCCTGGACGTCGAGTTCATGCAGCGACTCGGAGATAAGGTCAACATCATTCCGGTCATCGCTAAGGCCGATACAATGACGCCCGAAGAGTGCAAGGACTTCAAAGAACAGATAATGAAAGAAATCGGTCAGCACAAGATTAAAATTTACGACTTCCCGGAGAGCACGGGCGAGGAAGGGGACGGCGCTGACGTCACCAAGTCGCTGCGGGCGCGGGTACCCTTCGCCGTCGTCGGCGCCAACACCGTCATAGAGCAGGACGGGCGCCGCATACGGGGCAGGAAGTACCCTTGGGGCATCGCCGAGGTTGAAAATTTGGAGCACTGCGATTTTCTGGCGCTTCGGAACATGGTGATACGGACGCACCTTCAGGACCTGAAGGACGTGACCAGCTCTGTGCATTACGAGAACTATCGATGTCGCAAACTGGCCGGACTCTCGCACGACGGGAAGCCGCACAGAACCAACTCTAATAAGAATCCATTAGCACAGATGGAAGAGGAAAAACGCGAACACGACCTAAAGATGGAAAAAATGGAGAGTGAAATGGAACAAATATTCCAAACGAAAGTCCGCGACAAGCGCGCGAAGCTGAAGGAGTCCGAGGCGGAGCTGGCGCGGCGGCACGAGGCCACGCGGCGCGCGCTGGAGGCGCAGGCGCGCGAGCTGGACGAGCGGCAGCGCGCACTCAACGCCGAGCGCGCCGCCTGGGAGCGCGACACCGGCCTCTCGCTCGACGACCTGCGCAGGCGCTCGCTCGAGGCCAACAGCAAAGAAACCGTTGACGGCAAAGAAAAGAAGGACAAGAAAAAGAAAGATTGGTCTTTTAAGAGTGTCGCAACTGGAATATCTATATAA
Protein
MNGSTTTVNSGAVNGPGGTVNGIGAAPRTATLGTPSSYAYTSSGSLLGRGGSARPVPPPTLPKYSGSFSAGSTTGLREREREGTGGSHRLASLERLALRQRIIEQSANNQVASATIINSSSNGGLTSATVTLTDTATLQSKRELFFKSDTVSGGGVSASVPPSAAPPAPPSTLKDALSKRSATLPEPQETNHVEANGIATKHSNSVSVDVSKPPSTAPPPTPAPPVPISAPPAPVPTPLRSTENIMKKAEHPPVAPKPDLPKIEKPKTKELDGYVGFANLPNQVYRKAVKKGFEFTLMVVGESGLGKSTLINSLFLTEVYDKDKHPGPSLRVKKTVGVETSVVLLKENGVNLTLTIVDTPGFGDAVDNSNCWQPIIDFVESKYEEFLNAESRVARKAAPPDTRVHCCLYFIAPSGHGLKPLDVEFMQRLGDKVNIIPVIAKADTMTPEECKDFKEQIMKEIGQHKIKIYDFPESTGEEGDGADVTKSLRARVPFAVVGANTVIEQDGRRIRGRKYPWGIAEVENLEHCDFLALRNMVIRTHLQDLKDVTSSVHYENYRCRKLAGLSHDGKPHRTNSNKNPLAQMEEEKREHDLKMEKMESEMEQIFQTKVRDKRAKLKESEAELARRHEATRRALEAQARELDERQRALNAERAAWERDTGLSLDDLRRRSLEANSKETVDGKEKKDKKKKDWSFKSVATGISI
Summary
Similarity
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Uniprot
EMBL
Proteomes
Pfam
PF00735 Septin
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
PDB
2QAG
E-value=3.28976e-149,
Score=1357
Ontologies
GO
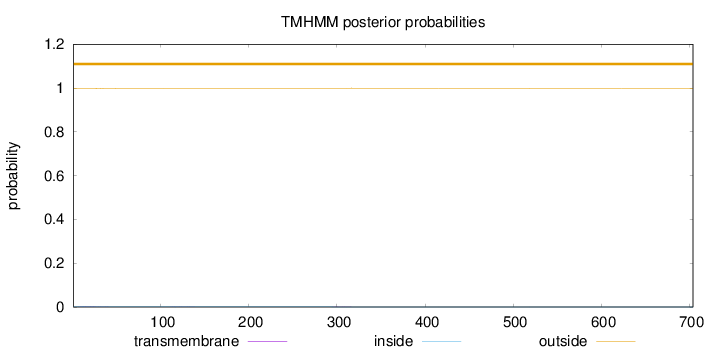
Topology
Length:
704
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06033
Exp number, first 60 AAs:
0.04246
Total prob of N-in:
0.00332
outside
1 - 704
Population Genetic Test Statistics
Pi
267.804959
Theta
200.865134
Tajima's D
1.120569
CLR
0.136264
CSRT
0.693415329233538
Interpretation
Uncertain
