Gene
KWMTBOMO02689
Pre Gene Modal
BGIBMGA003455
Annotation
PREDICTED:_alpha-catulin_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.2
Sequence
CDS
ATGGATATCCCAAAGGGAATGGCTCACCTTGAGATATCGAATAATTTTCTTCCAGGCCGCGCCATTGAACGAGTGTGTGCTGGGGCCGACGGAGCTGGCATGGTGTCCGCGGCAAGGGCGCTACTCGCGGCTGTCACCAGAGTCTTACTTTTGGCAGACATGGTAGTTGTTCGACAACTATTGCTTGCTAAGGATAAGGTGGCGCGATCGTTAGACCGGCTGGAGTCGGTCTCTAATTTCGCGGAGTTCGTGCGAGCATTCACCAAATTCGGCGGCGGTATGGTGGAACTGGCGCGGCTCACGGCCGAGCGCCGCGCCGACCTGCGGGACGAGCGACGGAGGGCCCAGGTCGCTGCTGCCAGGAATGTACTAGAACGTTCAACACTGATGCTCCTTACTTCGTCAAAGACATGCATGCGACATCCGGGAAGCGCATCGGCGAGAGAAAACCGTGACACTGTTTTTTGTCAAATGCGACGGGCGATGGACCTCATACACTACGTAGTACGAGACGGCTTGCCCGGCCACGAAGAACATGGACATGAGGCAGCTCAGTGGGAGGCAGGGACTGCATTGGGTGCTCTGCGAGGACTGTCATCCCAAGTAAGGGCAGCAAGAGCCCGGGGCGGAGCGGACGCCTCACGCAGACGAGCCCTAGCTGGCACGCTGCGGGCTTTGGTTGAACGAACGCACGACTTCACAGACTCCGCTTACACCTCGCATGAACACAGACAGCAAATACTAGCTCTGGCCGAGAGAATAGCTTACGAGCTGGAGAGACTCGTTGCCGTAGCGGTTTCGCTGGAGGAACAAGGTGGTATAAGTAGTAGCGCTGCACTGGAAAGTGCATGCACAGGCGCTACTGGTGCAGCATCAGAACTAGAACGAGCCCTAGTAGCCGCCGCTCGCGACCAAGCAAGAGACTTGGCCCCACTTGCAGACGAAGCTAAACGTCTCGCTTCGGACCTCGCACATATAGCCAGTTCCTGTGGCGAGAGAGAATCGGAGAGGTTGCATCATATAGCCAGTCGTCTCCACGAACAATTAGACCATATTATAGAGGTATGTAAACTACTTAGACACATAGCAATGTCGGAAACCCTTCAAGTCAGCGCAAAATTTGCCGAAATCAATCTAAGGATATACGGTCCTCAAGTGGTGACGGCAGCGCGCACACTAGCGGCATATCCAGGCAGTGCGGCCGCGCGGGAAAACCTTGACGTCTTCGTGGACATGTGGGCCTGGCTGCTCGCTGACGCCGCACGTCTCGCCACAGAACTGCTCCACATCACAGACGATAAACCCGATAAACAACAGTACAGCAGTTTACCAAGACCCGGTAAACACGGTACGACGAGTAAACCTTTAAAAGCCGTCAAATTGGACTCGGACGAGCAAGCAAAAATAGCGAAATCGGGTCTAGAAATGAAAATGATGTCTTCAGAAATGGACGCTGAGACTGAGAGATGGCAGGGCGCCGCCGCCGACGAAAACAACGACATCGTAAAAAGGGCAAAAAATATGTCTTCCATGGCTTTTGCTATGTATCAATTTACTAAAGGCGAGGGTCGTTTAAATACAACGCAAGACCTATTCACACAAGCAGAGTATTTTGCTGAAGAAGCGAATAGGCTCTATAAGATTGTAAGACAATTTTCATACCAGGTTCCTGCCGGTTCAACAAAAAAAGAACTTCTGGAACATCTGGACAAAGTACCGACCTACGTTCAACAATTACAATTTACGGTAAAGGAGCCCACGGTGGGACGCGCTGCTACATTCAGCAAAGTCGATAACGTCATCCAAGAGACCAAGCACCTCATGAACGTGATAAGTAAAGTTGTCACCACTTGCTTCGAGTGCGCGAACAAGTACAAATTGGAGTTCACGGGTTTATCAGCGGGTGGGGCTGGGTCTGGCGGGGGGCGAGGTACATCGCGGGATGAGGAGGGCGGTGGCGGAGGTGGTGGAGACGCCAAGCCGGGGGGGAGTAGCTCCGACACAGCCATGTGA
Protein
MDIPKGMAHLEISNNFLPGRAIERVCAGADGAGMVSAARALLAAVTRVLLLADMVVVRQLLLAKDKVARSLDRLESVSNFAEFVRAFTKFGGGMVELARLTAERRADLRDERRRAQVAAARNVLERSTLMLLTSSKTCMRHPGSASARENRDTVFCQMRRAMDLIHYVVRDGLPGHEEHGHEAAQWEAGTALGALRGLSSQVRAARARGGADASRRRALAGTLRALVERTHDFTDSAYTSHEHRQQILALAERIAYELERLVAVAVSLEEQGGISSSAALESACTGATGAASELERALVAAARDQARDLAPLADEAKRLASDLAHIASSCGERESERLHHIASRLHEQLDHIIEVCKLLRHIAMSETLQVSAKFAEINLRIYGPQVVTAARTLAAYPGSAAARENLDVFVDMWAWLLADAARLATELLHITDDKPDKQQYSSLPRPGKHGTTSKPLKAVKLDSDEQAKIAKSGLEMKMMSSEMDAETERWQGAAADENNDIVKRAKNMSSMAFAMYQFTKGEGRLNTTQDLFTQAEYFAEEANRLYKIVRQFSYQVPAGSTKKELLEHLDKVPTYVQQLQFTVKEPTVGRAATFSKVDNVIQETKHLMNVISKVVTTCFECANKYKLEFTGLSAGGAGSGGGRGTSRDEEGGGGGGGDAKPGGSSSDTAM
Summary
Uniprot
A0A2A4JKI0
A0A2A4JLD8
A0A212ESR6
A0A2H1W2Z1
A0A0L7LVK0
A0A194RL35
+ More
A0A194PPZ7 A0A0J7LA50 A0A151K0G0 A0A026VSZ5 E2B5J0 A0A158NMD3 F4W708 E2AZV3 A0A195C1U3 A0A195AX89 A0A151X7W8 A0A195DB05 K7IVA9 A0A310SLT4 A0A0L7QRJ2 A0A232F6X7 D6WHX5 A0A087ZXV1 A0A0M8ZWP4 A0A067RP86 N6TVN8 A0A1W4XIY7 A0A139WLL1 T1IXC7 A0A146M499 A0A0A9XDS0 A0A0A9XJ70 A0A0A9XGS7 A0A023F4F3 T1HZG9 A0A3Q0IW29 A0A154P371 A0A2A3EB84 E0VMD4 A0A2S2Q0D0 A0A0T6AYQ5 A0A2H8TWX9 E9GV61 A0A2J7QKU9 A0A2P8Z7D2 A0A2P2I2K4 A0A1S3JDR4 A0A1S3JE76 A0A1S3JF05 V9ILE5 A0A210R5Z0 F1KTV8 A0A0L8GDD3 A0A0V0S2D1 A0A3P6U816 A0A0V0XL88 A0A0V1AAE0 A0A0V1HJL9 A0A0V1CTA9 A0A0V0XBH6 A0A0V1MZM3 A0A0V0V130 A0A0V0UHR1 A0A0V1P2R8 A0A0V1BFM3 A0A0B2VCN7
A0A194PPZ7 A0A0J7LA50 A0A151K0G0 A0A026VSZ5 E2B5J0 A0A158NMD3 F4W708 E2AZV3 A0A195C1U3 A0A195AX89 A0A151X7W8 A0A195DB05 K7IVA9 A0A310SLT4 A0A0L7QRJ2 A0A232F6X7 D6WHX5 A0A087ZXV1 A0A0M8ZWP4 A0A067RP86 N6TVN8 A0A1W4XIY7 A0A139WLL1 T1IXC7 A0A146M499 A0A0A9XDS0 A0A0A9XJ70 A0A0A9XGS7 A0A023F4F3 T1HZG9 A0A3Q0IW29 A0A154P371 A0A2A3EB84 E0VMD4 A0A2S2Q0D0 A0A0T6AYQ5 A0A2H8TWX9 E9GV61 A0A2J7QKU9 A0A2P8Z7D2 A0A2P2I2K4 A0A1S3JDR4 A0A1S3JE76 A0A1S3JF05 V9ILE5 A0A210R5Z0 F1KTV8 A0A0L8GDD3 A0A0V0S2D1 A0A3P6U816 A0A0V0XL88 A0A0V1AAE0 A0A0V1HJL9 A0A0V1CTA9 A0A0V0XBH6 A0A0V1MZM3 A0A0V0V130 A0A0V0UHR1 A0A0V1P2R8 A0A0V1BFM3 A0A0B2VCN7
Pubmed
EMBL
NWSH01001105
PCG72585.1
PCG72586.1
AGBW02012767
OWR44494.1
ODYU01005960
+ More
SOQ47393.1 JTDY01000016 KOB79409.1 KQ460207 KPJ16691.1 KQ459597 KPI94809.1 LBMM01000074 KMR05066.1 KMR05067.1 KQ981253 KYN44181.1 KK108141 EZA46857.1 GL445834 EFN89034.1 ADTU01002236 ADTU01002237 ADTU01002238 ADTU01002239 ADTU01002240 ADTU01002241 GL887802 EGI69998.1 GL444277 EFN61082.1 KQ978344 KYM94817.1 KQ976716 KYM76801.1 KQ982431 KYQ56467.1 KQ981042 KYN10068.1 AAZX01001180 AAZX01011393 AAZX01012422 KQ764369 OAD54529.1 KQ414784 KOC61116.1 NNAY01000771 OXU26604.1 KQ971321 EFA00710.2 KQ435834 KOX71596.1 KK852549 KDR21554.1 APGK01052772 APGK01052773 APGK01052774 KB741216 ENN72461.1 KYB28715.1 JH431646 GDHC01004370 JAQ14259.1 GBHO01024732 GDHC01012629 JAG18872.1 JAQ06000.1 GBHO01024731 JAG18873.1 GBHO01024733 GDHC01018820 JAG18871.1 JAP99808.1 GBBI01002813 JAC15899.1 ACPB03016118 KQ434796 KZC05580.1 KZ288294 PBC28985.1 DS235307 EEB14540.1 GGMS01002003 MBY71206.1 LJIG01022513 KRT80186.1 GFXV01006624 MBW18429.1 GL732567 EFX76664.1 NEVH01013256 PNF29199.1 PYGN01000163 PSN52400.1 IACF01002581 LAB68229.1 JR053250 AEY61890.1 NEDP02000189 OWF56469.1 JI165785 ADY41312.1 KQ422385 KOF74953.1 JYDL01000047 KRX20629.1 UYRX01000128 VDK74918.1 JYDU01000225 JYDR01000047 JYDT01000117 JYDS01000286 JYDV01000012 KRX88719.1 KRY72237.1 KRY84312.1 KRZ18248.1 KRZ42850.1 JYDQ01000015 KRY21604.1 JYDP01000062 KRZ10226.1 JYDI01000104 KRY52491.1 JYDK01000002 KRX85382.1 JYDO01000021 KRZ77240.1 JYDN01000106 KRX57231.1 JYDJ01000002 KRX50966.1 JYDM01000055 KRZ90500.1 JYDH01000051 KRY35679.1 JPKZ01002041 KHN78740.1
SOQ47393.1 JTDY01000016 KOB79409.1 KQ460207 KPJ16691.1 KQ459597 KPI94809.1 LBMM01000074 KMR05066.1 KMR05067.1 KQ981253 KYN44181.1 KK108141 EZA46857.1 GL445834 EFN89034.1 ADTU01002236 ADTU01002237 ADTU01002238 ADTU01002239 ADTU01002240 ADTU01002241 GL887802 EGI69998.1 GL444277 EFN61082.1 KQ978344 KYM94817.1 KQ976716 KYM76801.1 KQ982431 KYQ56467.1 KQ981042 KYN10068.1 AAZX01001180 AAZX01011393 AAZX01012422 KQ764369 OAD54529.1 KQ414784 KOC61116.1 NNAY01000771 OXU26604.1 KQ971321 EFA00710.2 KQ435834 KOX71596.1 KK852549 KDR21554.1 APGK01052772 APGK01052773 APGK01052774 KB741216 ENN72461.1 KYB28715.1 JH431646 GDHC01004370 JAQ14259.1 GBHO01024732 GDHC01012629 JAG18872.1 JAQ06000.1 GBHO01024731 JAG18873.1 GBHO01024733 GDHC01018820 JAG18871.1 JAP99808.1 GBBI01002813 JAC15899.1 ACPB03016118 KQ434796 KZC05580.1 KZ288294 PBC28985.1 DS235307 EEB14540.1 GGMS01002003 MBY71206.1 LJIG01022513 KRT80186.1 GFXV01006624 MBW18429.1 GL732567 EFX76664.1 NEVH01013256 PNF29199.1 PYGN01000163 PSN52400.1 IACF01002581 LAB68229.1 JR053250 AEY61890.1 NEDP02000189 OWF56469.1 JI165785 ADY41312.1 KQ422385 KOF74953.1 JYDL01000047 KRX20629.1 UYRX01000128 VDK74918.1 JYDU01000225 JYDR01000047 JYDT01000117 JYDS01000286 JYDV01000012 KRX88719.1 KRY72237.1 KRY84312.1 KRZ18248.1 KRZ42850.1 JYDQ01000015 KRY21604.1 JYDP01000062 KRZ10226.1 JYDI01000104 KRY52491.1 JYDK01000002 KRX85382.1 JYDO01000021 KRZ77240.1 JYDN01000106 KRX57231.1 JYDJ01000002 KRX50966.1 JYDM01000055 KRZ90500.1 JYDH01000051 KRY35679.1 JPKZ01002041 KHN78740.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
UP000036403
+ More
UP000078541 UP000053097 UP000008237 UP000005205 UP000007755 UP000000311 UP000078542 UP000078540 UP000075809 UP000078492 UP000002358 UP000053825 UP000215335 UP000007266 UP000005203 UP000053105 UP000027135 UP000019118 UP000192223 UP000015103 UP000079169 UP000076502 UP000242457 UP000009046 UP000000305 UP000235965 UP000245037 UP000085678 UP000242188 UP000053454 UP000054630 UP000277928 UP000054632 UP000054805 UP000054815 UP000054826 UP000054995 UP000054783 UP000055024 UP000054653 UP000054673 UP000054843 UP000054681 UP000055048 UP000054924 UP000054776 UP000031036
UP000078541 UP000053097 UP000008237 UP000005205 UP000007755 UP000000311 UP000078542 UP000078540 UP000075809 UP000078492 UP000002358 UP000053825 UP000215335 UP000007266 UP000005203 UP000053105 UP000027135 UP000019118 UP000192223 UP000015103 UP000079169 UP000076502 UP000242457 UP000009046 UP000000305 UP000235965 UP000245037 UP000085678 UP000242188 UP000053454 UP000054630 UP000277928 UP000054632 UP000054805 UP000054815 UP000054826 UP000054995 UP000054783 UP000055024 UP000054653 UP000054673 UP000054843 UP000054681 UP000055048 UP000054924 UP000054776 UP000031036
Pfam
PF01044 Vinculin
Interpro
SUPFAM
SSF47220
SSF47220
ProteinModelPortal
A0A2A4JKI0
A0A2A4JLD8
A0A212ESR6
A0A2H1W2Z1
A0A0L7LVK0
A0A194RL35
+ More
A0A194PPZ7 A0A0J7LA50 A0A151K0G0 A0A026VSZ5 E2B5J0 A0A158NMD3 F4W708 E2AZV3 A0A195C1U3 A0A195AX89 A0A151X7W8 A0A195DB05 K7IVA9 A0A310SLT4 A0A0L7QRJ2 A0A232F6X7 D6WHX5 A0A087ZXV1 A0A0M8ZWP4 A0A067RP86 N6TVN8 A0A1W4XIY7 A0A139WLL1 T1IXC7 A0A146M499 A0A0A9XDS0 A0A0A9XJ70 A0A0A9XGS7 A0A023F4F3 T1HZG9 A0A3Q0IW29 A0A154P371 A0A2A3EB84 E0VMD4 A0A2S2Q0D0 A0A0T6AYQ5 A0A2H8TWX9 E9GV61 A0A2J7QKU9 A0A2P8Z7D2 A0A2P2I2K4 A0A1S3JDR4 A0A1S3JE76 A0A1S3JF05 V9ILE5 A0A210R5Z0 F1KTV8 A0A0L8GDD3 A0A0V0S2D1 A0A3P6U816 A0A0V0XL88 A0A0V1AAE0 A0A0V1HJL9 A0A0V1CTA9 A0A0V0XBH6 A0A0V1MZM3 A0A0V0V130 A0A0V0UHR1 A0A0V1P2R8 A0A0V1BFM3 A0A0B2VCN7
A0A194PPZ7 A0A0J7LA50 A0A151K0G0 A0A026VSZ5 E2B5J0 A0A158NMD3 F4W708 E2AZV3 A0A195C1U3 A0A195AX89 A0A151X7W8 A0A195DB05 K7IVA9 A0A310SLT4 A0A0L7QRJ2 A0A232F6X7 D6WHX5 A0A087ZXV1 A0A0M8ZWP4 A0A067RP86 N6TVN8 A0A1W4XIY7 A0A139WLL1 T1IXC7 A0A146M499 A0A0A9XDS0 A0A0A9XJ70 A0A0A9XGS7 A0A023F4F3 T1HZG9 A0A3Q0IW29 A0A154P371 A0A2A3EB84 E0VMD4 A0A2S2Q0D0 A0A0T6AYQ5 A0A2H8TWX9 E9GV61 A0A2J7QKU9 A0A2P8Z7D2 A0A2P2I2K4 A0A1S3JDR4 A0A1S3JE76 A0A1S3JF05 V9ILE5 A0A210R5Z0 F1KTV8 A0A0L8GDD3 A0A0V0S2D1 A0A3P6U816 A0A0V0XL88 A0A0V1AAE0 A0A0V1HJL9 A0A0V1CTA9 A0A0V0XBH6 A0A0V1MZM3 A0A0V0V130 A0A0V0UHR1 A0A0V1P2R8 A0A0V1BFM3 A0A0B2VCN7
PDB
6DV1
E-value=4.81004e-15,
Score=200
Ontologies
GO
PANTHER
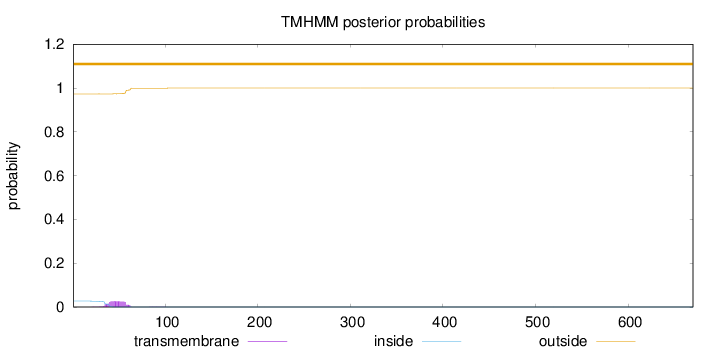
Topology
Length:
670
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.594289999999998
Exp number, first 60 AAs:
0.57139
Total prob of N-in:
0.02686
outside
1 - 670
Population Genetic Test Statistics
Pi
281.982244
Theta
194.985334
Tajima's D
1.325151
CLR
0.17379
CSRT
0.748912554372281
Interpretation
Uncertain
