Gene
KWMTBOMO02687
Pre Gene Modal
BGIBMGA003454
Annotation
PREDICTED:_myosin-IIIb-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.924
Sequence
CDS
ATGGCATATCGCGGGCTGTCCCAACATGTGGAACTAGACCGAGCTGCCGATCCTCGGGACAGATTCACGTTGCAGGAACTCATCGGCGAAGGCACATATGGCGAAGTATACTGTGCCAGAGATAAAAAGTCTGGTAGACGTGTAGCAGTCAAGATCTTGGAAAACATAGCTGAAAATATAGAAGAAATCGAAGAAGAGTTTCTAGTTTTTAGAGACTTGTCTAGTCATCCGAATATACCGGAGTTTTTTGGTTTGTTCTTGAAACGCGGCGCCAGCTCAGACGACGACCAAATATGGTTTGTGATGGAGCTGTGCACCGGTGGTTCGGTAACTGATTTATGTGCTGGAATGCGAGCCCGCGGTACAAATATTACTGAACCACAACTTGCATATGTGTTGCGGGGAACAGTTCGAGCACTGACTCACCTCCACGCTCACCGATGTATGCATCGTGATGTCAAAGGTCATAACATTCTTTTGACGGAAGATGCTGAGGTCAAACTCGTCGACTTTGGGGTTTCTTCACATTTGGCTGCTACAGCTGCGCGAAGAAATACTTCAGTCGGTACCCCATATTGGATGGCACCTGAGGTTATCGCATGTGAACAACAATTAGATGAGTCATACGACTGTAGATGTGATGTATGGTCAGTGGGTATAACCGCTATAGAACTGGCAGAAGGCGAACCTCCTCTATCGGGATTACATCCGATGCGAGCATTATTTCAAATACCAAGAAATCCACCACCCAGCTTGTCTCATCCAGAAATATTTTCGCCGCAACTTTCCGATTTTGTAGCAGACTGCTTAATAAAAGATATGAATCAAAGACCATTCGCGAGAGAGCTCTTGGAACATCCTCTGCTTCTTGCAGTTAGTAGCTTTGAAGACAAGATCCGGAAAGAGTTACAAGCTGAAATAAAACGACAGAGAGCAGAAGGACGAAGTTGTCGCGCTCCTGACGCAACTACAAAAAGGGGAAAATTGAAATCACACAGAAAAGCAAAACCTGAAAAAATGTATACTGACGACTTGGCAACGCTTGAAGTTTTAAGTGAAGATTCAATTGTTGAACAACTACAGAATAGATACAATCAAAATCAAATTTATACATATATTGGAGATATATTAGTGGCTGTTAATCCATTCACTGATATCGGCATTTATAACAGTAAGGTTTACCAAGGACGGTGTAGGTCGGATAATCCGCCTCACATCTTTGCCGTAGCTGATGCAGCCCATCAAGCACTAATGCATCAAAAACAAAACCAGGCCATAGTGATATCTGGCGAGAGTGGGGCTGGGAAGACTGAATCAGCAAATCTTTTATTAAAACATCTTGTCTTCCTGTCTAAGACTCAGAATCGTAATTTAGAAGAAAAAATTCTCCAAGTAAACCCAATAATGGAGGCATTCGGCAACGCTCGCACTGGAATAAACGCGAATAGTTCACGTTTCGGAAAATATCTAGATATGTCTATAATGGGAGTTGGAAGAATATCAGGAGCAAGGATATCAGTCTACCTTCTTGAGCAGTCTAGAGTTGTACATCAGGCTTTAGGTGAAAGCAATTTTCACGTGTTCTACTATCTGTATGATGGCTTAGAGAGCGAGGGCCGCTGGAGAAAGTTTTATCTAGATGAGCAACTAAAATCTAGACATCGTTATCTTCAGCCCTTATCTGTGGTTCATAGAGAGCACAATGTTCATCGTTGGCGACAGCTCAATCAAGCCTTTAAAGTAGTTGGATTTCTAGACGATGAAGTACAAATATTATATAAGATGCTCGCGGCAATATTACATCTTGGCGACATAGAGTTCGGTGAAACAGCTGGCGAAGATAATACTGATAACAGAGCAACTATAATAGACACTGCACCACTGCACAGAGCTTCTTGCCTATTGGGAGTTGAAACAGCAGATCTGCGTGAATGTTTAACAAGTAGCACAGTGGTCGCCCGAGGCGAAACTATAGCACGTCACAGTTCACCCACTGAAGCTGCCGTTGCCCGAGATGCTACAGCTCGTGGTCTATATGCAAGAGCATTCGATAGAATCGTCGAAAGAATTAACGCACTCCTCTCTCAAAACAGACACCGTCACGATCAACTCTCCATCGGCATTCTTGACATATTTGGGTTCGAAAACTTCAACAAAAATTCATTCGAGCAGCTCTGCATTAACATCGCCAACGAACAGATCCAGTATTACTTCAATCAACACATATTTACATGGGAGCAACAAGAATACATGGCTGAAGGTGTTCCAGTTGATCTCGTCGAATTTTCCGATAACAGACCTGTACTGGACATGTTACTGTCAAGGCCAATGGGCTTGTTAGCTCTGCTGGATGAAGAAAGTCGTTTCCCAAGAGCCACCGACAGAAGTCTTATTGAAAAGTTTCATAGTAATCTCAAAAGCAAATATTACGTTCGGCCAAAATCTGATGCAATTTGTTTTGCAATCCATCATTTTGCTGGTCGAGTCGTCTATCAGGCAGATGGGTTCTTGGAGAAGAATCGTAACTTTTTGCCACCTGAAGTCGTTCAGCTGATGCGACAGAGTCAATTCGATATCATACGGTTCTTATTTCAATGTCCAATCACGAAGACAGGAAATCTATATTCACCGCTACAAGGAGATATGGATAGAAGCTTGGAAAGTCCTTCTGGGGACACTCGAGATCGATTTAACAGTCGAGGACTTGCATCGCAATCACGAGCCCAGCAAACAGTCTCAACATACTTCCGATACTCGTTGATGGAGCTTCTACAGAAGATGGTGAGCGGCACTCCTCAGTTCGTGAGATGTCTAAAGCCGAACGACTCCAGATCTCCAAAGCACTTCGATCCGCCCAAAATACTAAAACAGCTTCGATACACTGGAGTATTAGAGACTATCAGGATAAGACAAAATGGTTTTTCCCATCGATTGAGTTTCGAAGAATTCTTGAAGCGATATGGCTTTTTGGCTTTTAGTCATGATGAAGAAATAAAGCCCAGTCGTGACACAAGTCGTCTACTGCTGCTCAGATTGAAGATGGACGGCTGGGCCCTCGGCAAATCCAAAGTGTTCCTCAAGTACTACCACGTTGAAAGACTCGCCAGAATCTACGAGGAACAGATAAGGAAAGTGATCCTGGTGCAAGCCTGCGTCAGGGGGTGGCTGGCTCGCCGCTACGTGGAGCGGCTGAAGGTACAGATGGCCATCTCGGTGCTGACCCTCCAGCGACACGTGCGCGGTTGGCTCACCAGAAAGCGAATCCGCGACCGACAGCGTCAGCTAGCCAGGGAGTTCCACGATGAGCAGATGTGCGACAGTCAGGGGATGCACAAATCTGGTTAG
Protein
MAYRGLSQHVELDRAADPRDRFTLQELIGEGTYGEVYCARDKKSGRRVAVKILENIAENIEEIEEEFLVFRDLSSHPNIPEFFGLFLKRGASSDDDQIWFVMELCTGGSVTDLCAGMRARGTNITEPQLAYVLRGTVRALTHLHAHRCMHRDVKGHNILLTEDAEVKLVDFGVSSHLAATAARRNTSVGTPYWMAPEVIACEQQLDESYDCRCDVWSVGITAIELAEGEPPLSGLHPMRALFQIPRNPPPSLSHPEIFSPQLSDFVADCLIKDMNQRPFARELLEHPLLLAVSSFEDKIRKELQAEIKRQRAEGRSCRAPDATTKRGKLKSHRKAKPEKMYTDDLATLEVLSEDSIVEQLQNRYNQNQIYTYIGDILVAVNPFTDIGIYNSKVYQGRCRSDNPPHIFAVADAAHQALMHQKQNQAIVISGESGAGKTESANLLLKHLVFLSKTQNRNLEEKILQVNPIMEAFGNARTGINANSSRFGKYLDMSIMGVGRISGARISVYLLEQSRVVHQALGESNFHVFYYLYDGLESEGRWRKFYLDEQLKSRHRYLQPLSVVHREHNVHRWRQLNQAFKVVGFLDDEVQILYKMLAAILHLGDIEFGETAGEDNTDNRATIIDTAPLHRASCLLGVETADLRECLTSSTVVARGETIARHSSPTEAAVARDATARGLYARAFDRIVERINALLSQNRHRHDQLSIGILDIFGFENFNKNSFEQLCINIANEQIQYYFNQHIFTWEQQEYMAEGVPVDLVEFSDNRPVLDMLLSRPMGLLALLDEESRFPRATDRSLIEKFHSNLKSKYYVRPKSDAICFAIHHFAGRVVYQADGFLEKNRNFLPPEVVQLMRQSQFDIIRFLFQCPITKTGNLYSPLQGDMDRSLESPSGDTRDRFNSRGLASQSRAQQTVSTYFRYSLMELLQKMVSGTPQFVRCLKPNDSRSPKHFDPPKILKQLRYTGVLETIRIRQNGFSHRLSFEEFLKRYGFLAFSHDEEIKPSRDTSRLLLLRLKMDGWALGKSKVFLKYYHVERLARIYEEQIRKVILVQACVRGWLARRYVERLKVQMAISVLTLQRHVRGWLTRKRIRDRQRQLAREFHDEQMCDSQGMHKSG
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9J1R7
A0A2A4JL97
A0A212ESM3
A0A194PNB2
A0A194RGW9
A0A195DB37
+ More
A0A182HW56 A0A084W8D1 A0A158NSE1 A0A195AXX9 A0A151K0S2 A0A182P5D9 A0A1S4FNX4 Q16U79 A0A0L7QRD4 Q16U80 A0A182X5U1 A0A1Y9GHR7 A0A1S4H5S4 A0A182WDQ6 A0A182FW15 A0A182YAW2 A0A182R6L5 A0A182Q3R7 A0A0M8ZUU7 B0WKI9 A0A195C3Q5 A0A182J2M0 E2AZV4 A0A310SLB6 A0A232F0C1 A0A1B0CRM0 A0A154P0W6 A0A336KEL3 A0A087ZXV0 A0A151X7W0 A0A0C9QB68 A0A0C9Q3V9 A0A182KKW2 A0A026VX80 A0A1B6H4M2 A0A1B6G5A1 A0A1B6GW40 A0A1B6KHC5 A0A1B6KVY8 A0A1B6KLX9 A0A1J1IY80 A0A1J1IW58 A0A1B6M0M8 A0A182K985 W5JSW7 A0A1B6CG25 A0A1B6DX50 A0A1B6DCZ0 E2B5J3 A0A182U731 A0A182M634 A0A2M4AKT5 A0A3L8D508 E0VH82 A0A2S2PD33 J9JL34 A0A146L935 A0A0P6GER5 E9GTF3 A0A0P5TRE8 A0A0P5HU10 A0A0P5SYQ6 A0A0P5I570 A0A0P5E4B9 A0A0P5XHN7 A0A0P4Z7K6 A0A0P5T762 A0A0P5VN02 A0A0P5Y2Y2 A0A0P5K3B6 A0A0P5J3U7 A0A0P4ZW12 A0A0P5EBH3 A0A0P5FE81 A0A0P5IMX7 A0A0P6D9E5 A0A0P6A5L2 A0A0P5R2B9 A0A0P5ZY02 A0A0P5DBL5 A0A0P6A2K4 A0A0P5DWI2 A0A0P5N0C9 A0A0P6GF05 A0A0P5DIA7 A0A0P6I3Q1 A0A0P6F7A7 A0A0P6GIT8 A0A0P5SS83 A0A0P5VN04 A0A0P5ZZ40 A0A0P4ZP84 A0A0P5WAW4 A0A0P4WSR3
A0A182HW56 A0A084W8D1 A0A158NSE1 A0A195AXX9 A0A151K0S2 A0A182P5D9 A0A1S4FNX4 Q16U79 A0A0L7QRD4 Q16U80 A0A182X5U1 A0A1Y9GHR7 A0A1S4H5S4 A0A182WDQ6 A0A182FW15 A0A182YAW2 A0A182R6L5 A0A182Q3R7 A0A0M8ZUU7 B0WKI9 A0A195C3Q5 A0A182J2M0 E2AZV4 A0A310SLB6 A0A232F0C1 A0A1B0CRM0 A0A154P0W6 A0A336KEL3 A0A087ZXV0 A0A151X7W0 A0A0C9QB68 A0A0C9Q3V9 A0A182KKW2 A0A026VX80 A0A1B6H4M2 A0A1B6G5A1 A0A1B6GW40 A0A1B6KHC5 A0A1B6KVY8 A0A1B6KLX9 A0A1J1IY80 A0A1J1IW58 A0A1B6M0M8 A0A182K985 W5JSW7 A0A1B6CG25 A0A1B6DX50 A0A1B6DCZ0 E2B5J3 A0A182U731 A0A182M634 A0A2M4AKT5 A0A3L8D508 E0VH82 A0A2S2PD33 J9JL34 A0A146L935 A0A0P6GER5 E9GTF3 A0A0P5TRE8 A0A0P5HU10 A0A0P5SYQ6 A0A0P5I570 A0A0P5E4B9 A0A0P5XHN7 A0A0P4Z7K6 A0A0P5T762 A0A0P5VN02 A0A0P5Y2Y2 A0A0P5K3B6 A0A0P5J3U7 A0A0P4ZW12 A0A0P5EBH3 A0A0P5FE81 A0A0P5IMX7 A0A0P6D9E5 A0A0P6A5L2 A0A0P5R2B9 A0A0P5ZY02 A0A0P5DBL5 A0A0P6A2K4 A0A0P5DWI2 A0A0P5N0C9 A0A0P6GF05 A0A0P5DIA7 A0A0P6I3Q1 A0A0P6F7A7 A0A0P6GIT8 A0A0P5SS83 A0A0P5VN04 A0A0P5ZZ40 A0A0P4ZP84 A0A0P5WAW4 A0A0P4WSR3
Pubmed
EMBL
BABH01008135
BABH01008136
NWSH01001105
PCG72589.1
AGBW02012767
OWR44495.1
+ More
KQ459597 KPI94807.1 KQ460207 KPJ16689.1 KQ981042 KYN10067.1 APCN01001469 ATLV01021413 KE525318 KFB46475.1 ADTU01002241 ADTU01002242 KQ976716 KYM76799.1 KQ981253 KYN44182.1 CH477630 EAT38082.1 KQ414784 KOC61114.1 EAT38083.1 AAAB01008816 AXCN02002140 KQ435834 KOX71595.1 DS231972 EDS29865.1 KQ978344 KYM94816.1 GL444277 EFN61083.1 KQ764369 OAD54527.1 NNAY01001436 OXU23980.1 AJWK01024934 AJWK01024935 KQ434796 KZC05579.1 UFQS01000325 UFQT01000325 SSX02881.1 SSX23248.1 KQ982431 KYQ56466.1 GBYB01011693 JAG81460.1 GBYB01008743 JAG78510.1 KK107642 EZA48367.1 GECZ01000124 JAS69645.1 GECZ01012149 JAS57620.1 GECZ01003115 JAS66654.1 GEBQ01029127 JAT10850.1 GEBQ01024351 JAT15626.1 GEBQ01027512 JAT12465.1 CVRI01000063 CRL04516.1 CRL04515.1 GEBQ01010499 JAT29478.1 ADMH02000252 ETN67236.1 GEDC01024949 JAS12349.1 GEDC01007040 JAS30258.1 GEDC01013735 JAS23563.1 GL445834 EFN89037.1 AXCM01006247 GGFK01007907 MBW41228.1 QOIP01000013 RLU15535.1 DS235161 EEB12738.1 GGMR01014730 MBY27349.1 ABLF02014987 ABLF02014988 GDHC01014893 JAQ03736.1 GDIQ01034146 JAN60591.1 GL732564 EFX77076.1 GDIP01123817 JAL79897.1 GDIQ01223334 JAK28391.1 GDIQ01098567 JAL53159.1 GDIQ01223335 JAK28390.1 GDIP01165262 JAJ58140.1 GDIP01073784 LRGB01000024 JAM29931.1 KZS21454.1 GDIP01217510 JAJ05892.1 GDIP01130022 JAL73692.1 GDIP01097822 JAM05893.1 GDIP01064683 JAM39032.1 GDIQ01215888 JAK35837.1 GDIQ01203584 JAK48141.1 GDIP01220759 JAJ02643.1 GDIP01148868 JAJ74534.1 GDIP01148869 JAJ74533.1 GDIQ01211930 JAK39795.1 GDIQ01081490 JAN13247.1 GDIP01034886 JAM68829.1 GDIQ01106469 JAL45257.1 GDIP01037086 JAM66629.1 GDIP01158472 JAJ64930.1 GDIP01035933 JAM67782.1 GDIP01167868 JAJ55534.1 GDIQ01170942 JAK80783.1 GDIQ01034145 JAN60592.1 GDIP01159272 JAJ64130.1 GDIQ01035449 JAN59288.1 GDIQ01064488 GDIQ01059045 JAN30249.1 GDIQ01032656 JAN62081.1 GDIQ01091519 JAL60207.1 GDIP01097821 JAM05894.1 GDIP01037085 JAM66630.1 GDIP01209910 JAJ13492.1 GDIP01088682 JAM15033.1 GDIP01253651 JAI69750.1
KQ459597 KPI94807.1 KQ460207 KPJ16689.1 KQ981042 KYN10067.1 APCN01001469 ATLV01021413 KE525318 KFB46475.1 ADTU01002241 ADTU01002242 KQ976716 KYM76799.1 KQ981253 KYN44182.1 CH477630 EAT38082.1 KQ414784 KOC61114.1 EAT38083.1 AAAB01008816 AXCN02002140 KQ435834 KOX71595.1 DS231972 EDS29865.1 KQ978344 KYM94816.1 GL444277 EFN61083.1 KQ764369 OAD54527.1 NNAY01001436 OXU23980.1 AJWK01024934 AJWK01024935 KQ434796 KZC05579.1 UFQS01000325 UFQT01000325 SSX02881.1 SSX23248.1 KQ982431 KYQ56466.1 GBYB01011693 JAG81460.1 GBYB01008743 JAG78510.1 KK107642 EZA48367.1 GECZ01000124 JAS69645.1 GECZ01012149 JAS57620.1 GECZ01003115 JAS66654.1 GEBQ01029127 JAT10850.1 GEBQ01024351 JAT15626.1 GEBQ01027512 JAT12465.1 CVRI01000063 CRL04516.1 CRL04515.1 GEBQ01010499 JAT29478.1 ADMH02000252 ETN67236.1 GEDC01024949 JAS12349.1 GEDC01007040 JAS30258.1 GEDC01013735 JAS23563.1 GL445834 EFN89037.1 AXCM01006247 GGFK01007907 MBW41228.1 QOIP01000013 RLU15535.1 DS235161 EEB12738.1 GGMR01014730 MBY27349.1 ABLF02014987 ABLF02014988 GDHC01014893 JAQ03736.1 GDIQ01034146 JAN60591.1 GL732564 EFX77076.1 GDIP01123817 JAL79897.1 GDIQ01223334 JAK28391.1 GDIQ01098567 JAL53159.1 GDIQ01223335 JAK28390.1 GDIP01165262 JAJ58140.1 GDIP01073784 LRGB01000024 JAM29931.1 KZS21454.1 GDIP01217510 JAJ05892.1 GDIP01130022 JAL73692.1 GDIP01097822 JAM05893.1 GDIP01064683 JAM39032.1 GDIQ01215888 JAK35837.1 GDIQ01203584 JAK48141.1 GDIP01220759 JAJ02643.1 GDIP01148868 JAJ74534.1 GDIP01148869 JAJ74533.1 GDIQ01211930 JAK39795.1 GDIQ01081490 JAN13247.1 GDIP01034886 JAM68829.1 GDIQ01106469 JAL45257.1 GDIP01037086 JAM66629.1 GDIP01158472 JAJ64930.1 GDIP01035933 JAM67782.1 GDIP01167868 JAJ55534.1 GDIQ01170942 JAK80783.1 GDIQ01034145 JAN60592.1 GDIP01159272 JAJ64130.1 GDIQ01035449 JAN59288.1 GDIQ01064488 GDIQ01059045 JAN30249.1 GDIQ01032656 JAN62081.1 GDIQ01091519 JAL60207.1 GDIP01097821 JAM05894.1 GDIP01037085 JAM66630.1 GDIP01209910 JAJ13492.1 GDIP01088682 JAM15033.1 GDIP01253651 JAI69750.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000078492
+ More
UP000075840 UP000030765 UP000005205 UP000078540 UP000078541 UP000075885 UP000008820 UP000053825 UP000076407 UP000075920 UP000069272 UP000076408 UP000075900 UP000075886 UP000053105 UP000002320 UP000078542 UP000075880 UP000000311 UP000215335 UP000092461 UP000076502 UP000005203 UP000075809 UP000075882 UP000053097 UP000183832 UP000075881 UP000000673 UP000008237 UP000075902 UP000075883 UP000279307 UP000009046 UP000007819 UP000000305 UP000076858
UP000075840 UP000030765 UP000005205 UP000078540 UP000078541 UP000075885 UP000008820 UP000053825 UP000076407 UP000075920 UP000069272 UP000076408 UP000075900 UP000075886 UP000053105 UP000002320 UP000078542 UP000075880 UP000000311 UP000215335 UP000092461 UP000076502 UP000005203 UP000075809 UP000075882 UP000053097 UP000183832 UP000075881 UP000000673 UP000008237 UP000075902 UP000075883 UP000279307 UP000009046 UP000007819 UP000000305 UP000076858
PRIDE
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR008271 Ser/Thr_kinase_AS
IPR027417 P-loop_NTPase
IPR001609 Myosin_head_motor_dom
IPR017441 Protein_kinase_ATP_BS
IPR036723 Alpha-catenin/vinculin-like_sf
IPR036961 Kinesin_motor_dom_sf
IPR000719 Prot_kinase_dom
IPR000048 IQ_motif_EF-hand-BS
IPR036083 MYSc_Myo3
IPR006077 Vinculin/catenin
IPR041426 Mos1_HTH
IPR008271 Ser/Thr_kinase_AS
IPR027417 P-loop_NTPase
IPR001609 Myosin_head_motor_dom
IPR017441 Protein_kinase_ATP_BS
IPR036723 Alpha-catenin/vinculin-like_sf
IPR036961 Kinesin_motor_dom_sf
IPR000719 Prot_kinase_dom
IPR000048 IQ_motif_EF-hand-BS
IPR036083 MYSc_Myo3
IPR006077 Vinculin/catenin
IPR041426 Mos1_HTH
Gene 3D
ProteinModelPortal
H9J1R7
A0A2A4JL97
A0A212ESM3
A0A194PNB2
A0A194RGW9
A0A195DB37
+ More
A0A182HW56 A0A084W8D1 A0A158NSE1 A0A195AXX9 A0A151K0S2 A0A182P5D9 A0A1S4FNX4 Q16U79 A0A0L7QRD4 Q16U80 A0A182X5U1 A0A1Y9GHR7 A0A1S4H5S4 A0A182WDQ6 A0A182FW15 A0A182YAW2 A0A182R6L5 A0A182Q3R7 A0A0M8ZUU7 B0WKI9 A0A195C3Q5 A0A182J2M0 E2AZV4 A0A310SLB6 A0A232F0C1 A0A1B0CRM0 A0A154P0W6 A0A336KEL3 A0A087ZXV0 A0A151X7W0 A0A0C9QB68 A0A0C9Q3V9 A0A182KKW2 A0A026VX80 A0A1B6H4M2 A0A1B6G5A1 A0A1B6GW40 A0A1B6KHC5 A0A1B6KVY8 A0A1B6KLX9 A0A1J1IY80 A0A1J1IW58 A0A1B6M0M8 A0A182K985 W5JSW7 A0A1B6CG25 A0A1B6DX50 A0A1B6DCZ0 E2B5J3 A0A182U731 A0A182M634 A0A2M4AKT5 A0A3L8D508 E0VH82 A0A2S2PD33 J9JL34 A0A146L935 A0A0P6GER5 E9GTF3 A0A0P5TRE8 A0A0P5HU10 A0A0P5SYQ6 A0A0P5I570 A0A0P5E4B9 A0A0P5XHN7 A0A0P4Z7K6 A0A0P5T762 A0A0P5VN02 A0A0P5Y2Y2 A0A0P5K3B6 A0A0P5J3U7 A0A0P4ZW12 A0A0P5EBH3 A0A0P5FE81 A0A0P5IMX7 A0A0P6D9E5 A0A0P6A5L2 A0A0P5R2B9 A0A0P5ZY02 A0A0P5DBL5 A0A0P6A2K4 A0A0P5DWI2 A0A0P5N0C9 A0A0P6GF05 A0A0P5DIA7 A0A0P6I3Q1 A0A0P6F7A7 A0A0P6GIT8 A0A0P5SS83 A0A0P5VN04 A0A0P5ZZ40 A0A0P4ZP84 A0A0P5WAW4 A0A0P4WSR3
A0A182HW56 A0A084W8D1 A0A158NSE1 A0A195AXX9 A0A151K0S2 A0A182P5D9 A0A1S4FNX4 Q16U79 A0A0L7QRD4 Q16U80 A0A182X5U1 A0A1Y9GHR7 A0A1S4H5S4 A0A182WDQ6 A0A182FW15 A0A182YAW2 A0A182R6L5 A0A182Q3R7 A0A0M8ZUU7 B0WKI9 A0A195C3Q5 A0A182J2M0 E2AZV4 A0A310SLB6 A0A232F0C1 A0A1B0CRM0 A0A154P0W6 A0A336KEL3 A0A087ZXV0 A0A151X7W0 A0A0C9QB68 A0A0C9Q3V9 A0A182KKW2 A0A026VX80 A0A1B6H4M2 A0A1B6G5A1 A0A1B6GW40 A0A1B6KHC5 A0A1B6KVY8 A0A1B6KLX9 A0A1J1IY80 A0A1J1IW58 A0A1B6M0M8 A0A182K985 W5JSW7 A0A1B6CG25 A0A1B6DX50 A0A1B6DCZ0 E2B5J3 A0A182U731 A0A182M634 A0A2M4AKT5 A0A3L8D508 E0VH82 A0A2S2PD33 J9JL34 A0A146L935 A0A0P6GER5 E9GTF3 A0A0P5TRE8 A0A0P5HU10 A0A0P5SYQ6 A0A0P5I570 A0A0P5E4B9 A0A0P5XHN7 A0A0P4Z7K6 A0A0P5T762 A0A0P5VN02 A0A0P5Y2Y2 A0A0P5K3B6 A0A0P5J3U7 A0A0P4ZW12 A0A0P5EBH3 A0A0P5FE81 A0A0P5IMX7 A0A0P6D9E5 A0A0P6A5L2 A0A0P5R2B9 A0A0P5ZY02 A0A0P5DBL5 A0A0P6A2K4 A0A0P5DWI2 A0A0P5N0C9 A0A0P6GF05 A0A0P5DIA7 A0A0P6I3Q1 A0A0P6F7A7 A0A0P6GIT8 A0A0P5SS83 A0A0P5VN04 A0A0P5ZZ40 A0A0P4ZP84 A0A0P5WAW4 A0A0P4WSR3
PDB
5I0I
E-value=8.42592e-136,
Score=1243
Ontologies
GO
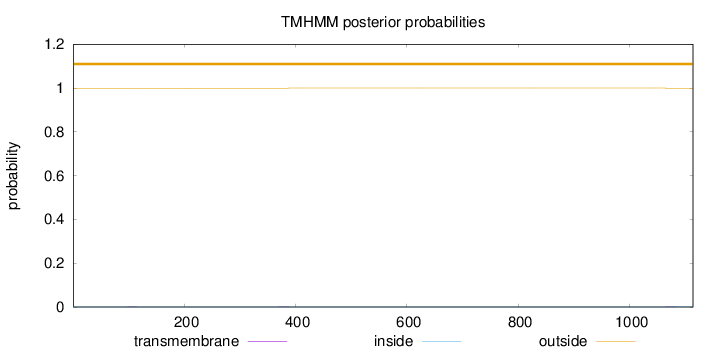
Topology
Length:
1114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02536
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00055
outside
1 - 1114
Population Genetic Test Statistics
Pi
195.152884
Theta
217.595329
Tajima's D
-1.356733
CLR
1245.543811
CSRT
0.0819459027048648
Interpretation
Uncertain
