Gene
KWMTBOMO02675
Pre Gene Modal
BGIBMGA003451
Annotation
guanylate_kinase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.257
Sequence
CDS
ATGGTTCAGAAAGGGCCTCGTCCTCTGGTACTGTGCGGACCTTCAGGCTCAGGCAAGAGCACGTTGTTGAAGCGGCTGCTGAAAGAATTTCCCGACAAGTTTGGCTTCAGTGTCTCCCATACAACAAGAGCACCACGGGCTGGTGAAAAAAACGGAGTGCATTATCATTTCACTAACCTAAACGATATGTCAACTGCTATTGAAAAAGGTGAATTTATAGAAACTGCAATTTTCAGTGGAAACATTTATGGAACAAGCAAGAAAGCAGTAGATGATGTACGTCGTACTGGCAAAATATGTGTGCTGGATATTGAAATGGAAGGGGTGAAGCAAATCAAACGCACAGACCTGGATCCCCTGTTGGTCTTTGTGATGCCACCATCTATAGATGAACTGGAACGAAGACTCCGCGCCCGTAATACGGAACAAGAAGATAGCTTGAAAAAGAGATTGGAAACGGCACGAAGAGAGATTAAATTTGGTCAAGAACCGGGAAACTTCAATATCATCATTTTGAATGATAATTTGGACAAAGCCTATAATGAGCTGCGTGATTTTATTACACAAAATATTGAAGACAATGATCAAGATGACACCAATGAGAAAACTACAAACAACAAGTGA
Protein
MVQKGPRPLVLCGPSGSGKSTLLKRLLKEFPDKFGFSVSHTTRAPRAGEKNGVHYHFTNLNDMSTAIEKGEFIETAIFSGNIYGTSKKAVDDVRRTGKICVLDIEMEGVKQIKRTDLDPLLVFVMPPSIDELERRLRARNTEQEDSLKKRLETARREIKFGQEPGNFNIIILNDNLDKAYNELRDFITQNIEDNDQDDTNEKTTNNK
Summary
Uniprot
Q2F688
H9J1R4
A0A2A4J0T4
A0A194PP90
S4P8U2
A0A212F5A1
+ More
A0A2H1WW91 A0A194RH82 A0A182MEB2 A0A336M2W3 A0A336LZ35 A0A182P5E5 A0A182R6M1 A0A182SDB9 A0A182WDP9 A0A1Y9GZ48 T1EAE3 W5JW57 A0A182YAW9 A0A182FW20 A0A2M3Z716 A0A2M4AJ97 A0A182N251 A0A182QHB0 A0A182HW61 A0A182JP11 A0A182VP56 Q16RC6 A0A182X5T4 Q7PSP6 A0A182KKX1 A0A1L8DLB9 A0A2M4CHY8 A0A182UE94 U5ETQ0 A0A084W8C1 A0A1L8DL95 A0A1B0GIR7 A0A2M4CGY4 A0A0K8TNB1 T1E2Z8 A0A1Q3F9J0 A0A182G5S9 A0A182IJS1 A0A023EN27 A0A1Q3F865 A0A1Q3G294 B0WKH0 A0A1Q3F7A6 A0A1Q3G251 A0A0L7LKH9 A0A0N0BJA3 A0A232EYV2 A0A0L7R1U6 E2A778 E9IML8 A0A088A648 A0A151I6R0 A0A151WP82 V9IE02 A0A151K112 A0A2A3EIZ8 A0A0J7KTJ1 T1D4M7 A0A151IV44 A0A195BAI1 K7J441 E2BHJ8 A0A1B6I671 A0A158NUV1 A0A1B6JPA2 A0A026W458 A0A1B6MG04 A0A1B6L1T3 A0A1B6G1E6 A0A0M4ENL2 A0A1B6MVC5 A0A067QSJ5 B3GC12 B3GC07 B3GBZ2 B3GC09 B3GBZ1 B4L9G8 A0A0T6AZA7 B4LB54 B4PEG7 E0VGF1 B3GBZ9 A0A1I8MEM7 Q29E07 T1PE16 A0A1I8P2T3 A0A0J9UIP9 B3M5K5 A0A1I8P2N4 A0A3B0JMK7 B4HDR1 B4H437 B4J0Y2
A0A2H1WW91 A0A194RH82 A0A182MEB2 A0A336M2W3 A0A336LZ35 A0A182P5E5 A0A182R6M1 A0A182SDB9 A0A182WDP9 A0A1Y9GZ48 T1EAE3 W5JW57 A0A182YAW9 A0A182FW20 A0A2M3Z716 A0A2M4AJ97 A0A182N251 A0A182QHB0 A0A182HW61 A0A182JP11 A0A182VP56 Q16RC6 A0A182X5T4 Q7PSP6 A0A182KKX1 A0A1L8DLB9 A0A2M4CHY8 A0A182UE94 U5ETQ0 A0A084W8C1 A0A1L8DL95 A0A1B0GIR7 A0A2M4CGY4 A0A0K8TNB1 T1E2Z8 A0A1Q3F9J0 A0A182G5S9 A0A182IJS1 A0A023EN27 A0A1Q3F865 A0A1Q3G294 B0WKH0 A0A1Q3F7A6 A0A1Q3G251 A0A0L7LKH9 A0A0N0BJA3 A0A232EYV2 A0A0L7R1U6 E2A778 E9IML8 A0A088A648 A0A151I6R0 A0A151WP82 V9IE02 A0A151K112 A0A2A3EIZ8 A0A0J7KTJ1 T1D4M7 A0A151IV44 A0A195BAI1 K7J441 E2BHJ8 A0A1B6I671 A0A158NUV1 A0A1B6JPA2 A0A026W458 A0A1B6MG04 A0A1B6L1T3 A0A1B6G1E6 A0A0M4ENL2 A0A1B6MVC5 A0A067QSJ5 B3GC12 B3GC07 B3GBZ2 B3GC09 B3GBZ1 B4L9G8 A0A0T6AZA7 B4LB54 B4PEG7 E0VGF1 B3GBZ9 A0A1I8MEM7 Q29E07 T1PE16 A0A1I8P2T3 A0A0J9UIP9 B3M5K5 A0A1I8P2N4 A0A3B0JMK7 B4HDR1 B4H437 B4J0Y2
Pubmed
EMBL
DQ311184
ABD36129.1
BABH01008148
BABH01008149
NWSH01003921
PCG65685.1
+ More
KQ459597 KPI94798.1 GAIX01004039 JAA88521.1 AGBW02010215 OWR48916.1 ODYU01011535 SOQ57339.1 KQ460207 KPJ16680.1 AXCM01001638 UFQS01000325 UFQT01000325 SSX02876.1 SSX23243.1 SSX02877.1 SSX23244.1 GAMD01000507 JAB01084.1 ADMH02000211 ETN67365.1 GGFM01003572 MBW24323.1 GGFK01007534 MBW40855.1 AXCN02001743 APCN01001469 CH477717 EAT36958.1 AAAB01008816 EAA05230.5 GFDF01006842 JAV07242.1 GGFL01000667 MBW64845.1 GANO01001804 JAB58067.1 ATLV01021410 KE525318 KFB46465.1 GFDF01006841 JAV07243.1 AJWK01015345 GGFL01000429 MBW64607.1 GDAI01002012 JAI15591.1 GALA01000835 JAA94017.1 GFDL01010843 JAV24202.1 JXUM01044193 JXUM01044194 JXUM01044195 KQ561390 KXJ78737.1 GAPW01003313 JAC10285.1 GFDL01011280 JAV23765.1 GFDL01001097 JAV33948.1 DS231972 EDS29846.1 GFDL01011594 JAV23451.1 GFDL01001165 JAV33880.1 JTDY01000769 KOB75940.1 KQ435719 KOX78842.1 NNAY01001595 OXU23480.1 KQ414667 KOC64814.1 GL437267 EFN70734.1 GL764212 EFZ18185.1 KQ978473 KYM93705.1 KQ982905 KYQ49505.1 JR039877 JR039878 AEY58891.1 AEY58892.1 KQ981296 KYN43086.1 KZ288229 PBC31697.1 LBMM01003344 KMQ93631.1 GALA01000888 JAA93964.1 KQ980926 KYN11382.1 KQ976532 KYM81556.1 AAZX01004540 GL448301 EFN84851.1 GECU01025270 GECU01015259 JAS82436.1 JAS92447.1 ADTU01026686 GECU01033044 GECU01006721 JAS74662.1 JAT00986.1 KK107435 EZA50870.1 GEBQ01005097 JAT34880.1 GEBQ01022310 JAT17667.1 GECZ01013527 JAS56242.1 CP012525 ALC43555.1 GEBQ01030928 GEBQ01029715 GEBQ01000089 JAT09049.1 JAT10262.1 JAT39888.1 KK853332 KDR08382.1 EU669693 ACD69451.1 EU669688 EU669689 EU669691 EU669692 EU669695 EU669697 EU669699 EU669700 EU669701 EU669702 EU669703 ACD69446.1 ACD69447.1 ACD69449.1 ACD69450.1 ACD69453.1 ACD69455.1 ACD69457.1 ACD69458.1 ACD69459.1 ACD69460.1 ACD69461.1 EU669673 EU669674 EU669675 EU669679 EU669682 EU669683 EU669685 ACD69431.1 ACD69432.1 ACD69433.1 ACD69437.1 ACD69440.1 ACD69441.1 ACD69443.1 EU669690 EU669694 EU669696 EU669698 ACD69448.1 ACD69452.1 ACD69454.1 ACD69456.1 EU669672 EU669676 EU669677 EU669678 EU669684 EU669686 EU669687 ACD69430.1 ACD69434.1 ACD69435.1 ACD69436.1 ACD69442.1 ACD69444.1 ACD69445.1 CH933816 EDW17343.1 LJIG01022451 KRT80527.1 CH940647 EDW68618.1 CM000159 EDW94033.1 DS235140 EEB12457.1 EU669680 EU669681 ACD69438.1 ACD69439.1 CH379070 EAL30256.1 KA646193 AFP60822.1 CM002912 KMY98870.1 CH902618 EDV40639.1 OUUW01000002 SPP76710.1 CH480815 EDW41001.1 CH479208 EDW31152.1 CH916366 EDV95803.1
KQ459597 KPI94798.1 GAIX01004039 JAA88521.1 AGBW02010215 OWR48916.1 ODYU01011535 SOQ57339.1 KQ460207 KPJ16680.1 AXCM01001638 UFQS01000325 UFQT01000325 SSX02876.1 SSX23243.1 SSX02877.1 SSX23244.1 GAMD01000507 JAB01084.1 ADMH02000211 ETN67365.1 GGFM01003572 MBW24323.1 GGFK01007534 MBW40855.1 AXCN02001743 APCN01001469 CH477717 EAT36958.1 AAAB01008816 EAA05230.5 GFDF01006842 JAV07242.1 GGFL01000667 MBW64845.1 GANO01001804 JAB58067.1 ATLV01021410 KE525318 KFB46465.1 GFDF01006841 JAV07243.1 AJWK01015345 GGFL01000429 MBW64607.1 GDAI01002012 JAI15591.1 GALA01000835 JAA94017.1 GFDL01010843 JAV24202.1 JXUM01044193 JXUM01044194 JXUM01044195 KQ561390 KXJ78737.1 GAPW01003313 JAC10285.1 GFDL01011280 JAV23765.1 GFDL01001097 JAV33948.1 DS231972 EDS29846.1 GFDL01011594 JAV23451.1 GFDL01001165 JAV33880.1 JTDY01000769 KOB75940.1 KQ435719 KOX78842.1 NNAY01001595 OXU23480.1 KQ414667 KOC64814.1 GL437267 EFN70734.1 GL764212 EFZ18185.1 KQ978473 KYM93705.1 KQ982905 KYQ49505.1 JR039877 JR039878 AEY58891.1 AEY58892.1 KQ981296 KYN43086.1 KZ288229 PBC31697.1 LBMM01003344 KMQ93631.1 GALA01000888 JAA93964.1 KQ980926 KYN11382.1 KQ976532 KYM81556.1 AAZX01004540 GL448301 EFN84851.1 GECU01025270 GECU01015259 JAS82436.1 JAS92447.1 ADTU01026686 GECU01033044 GECU01006721 JAS74662.1 JAT00986.1 KK107435 EZA50870.1 GEBQ01005097 JAT34880.1 GEBQ01022310 JAT17667.1 GECZ01013527 JAS56242.1 CP012525 ALC43555.1 GEBQ01030928 GEBQ01029715 GEBQ01000089 JAT09049.1 JAT10262.1 JAT39888.1 KK853332 KDR08382.1 EU669693 ACD69451.1 EU669688 EU669689 EU669691 EU669692 EU669695 EU669697 EU669699 EU669700 EU669701 EU669702 EU669703 ACD69446.1 ACD69447.1 ACD69449.1 ACD69450.1 ACD69453.1 ACD69455.1 ACD69457.1 ACD69458.1 ACD69459.1 ACD69460.1 ACD69461.1 EU669673 EU669674 EU669675 EU669679 EU669682 EU669683 EU669685 ACD69431.1 ACD69432.1 ACD69433.1 ACD69437.1 ACD69440.1 ACD69441.1 ACD69443.1 EU669690 EU669694 EU669696 EU669698 ACD69448.1 ACD69452.1 ACD69454.1 ACD69456.1 EU669672 EU669676 EU669677 EU669678 EU669684 EU669686 EU669687 ACD69430.1 ACD69434.1 ACD69435.1 ACD69436.1 ACD69442.1 ACD69444.1 ACD69445.1 CH933816 EDW17343.1 LJIG01022451 KRT80527.1 CH940647 EDW68618.1 CM000159 EDW94033.1 DS235140 EEB12457.1 EU669680 EU669681 ACD69438.1 ACD69439.1 CH379070 EAL30256.1 KA646193 AFP60822.1 CM002912 KMY98870.1 CH902618 EDV40639.1 OUUW01000002 SPP76710.1 CH480815 EDW41001.1 CH479208 EDW31152.1 CH916366 EDV95803.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000075883
+ More
UP000075885 UP000075900 UP000075901 UP000075920 UP000075884 UP000000673 UP000076408 UP000069272 UP000075886 UP000075840 UP000075881 UP000075903 UP000008820 UP000076407 UP000007062 UP000075882 UP000075902 UP000030765 UP000092461 UP000069940 UP000249989 UP000075880 UP000002320 UP000037510 UP000053105 UP000215335 UP000053825 UP000000311 UP000005203 UP000078542 UP000075809 UP000078541 UP000242457 UP000036403 UP000078492 UP000078540 UP000002358 UP000008237 UP000005205 UP000053097 UP000092553 UP000027135 UP000009192 UP000008792 UP000002282 UP000009046 UP000095301 UP000001819 UP000095300 UP000007801 UP000268350 UP000001292 UP000008744 UP000001070
UP000075885 UP000075900 UP000075901 UP000075920 UP000075884 UP000000673 UP000076408 UP000069272 UP000075886 UP000075840 UP000075881 UP000075903 UP000008820 UP000076407 UP000007062 UP000075882 UP000075902 UP000030765 UP000092461 UP000069940 UP000249989 UP000075880 UP000002320 UP000037510 UP000053105 UP000215335 UP000053825 UP000000311 UP000005203 UP000078542 UP000075809 UP000078541 UP000242457 UP000036403 UP000078492 UP000078540 UP000002358 UP000008237 UP000005205 UP000053097 UP000092553 UP000027135 UP000009192 UP000008792 UP000002282 UP000009046 UP000095301 UP000001819 UP000095300 UP000007801 UP000268350 UP000001292 UP000008744 UP000001070
Pfam
PF00625 Guanylate_kin
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q2F688
H9J1R4
A0A2A4J0T4
A0A194PP90
S4P8U2
A0A212F5A1
+ More
A0A2H1WW91 A0A194RH82 A0A182MEB2 A0A336M2W3 A0A336LZ35 A0A182P5E5 A0A182R6M1 A0A182SDB9 A0A182WDP9 A0A1Y9GZ48 T1EAE3 W5JW57 A0A182YAW9 A0A182FW20 A0A2M3Z716 A0A2M4AJ97 A0A182N251 A0A182QHB0 A0A182HW61 A0A182JP11 A0A182VP56 Q16RC6 A0A182X5T4 Q7PSP6 A0A182KKX1 A0A1L8DLB9 A0A2M4CHY8 A0A182UE94 U5ETQ0 A0A084W8C1 A0A1L8DL95 A0A1B0GIR7 A0A2M4CGY4 A0A0K8TNB1 T1E2Z8 A0A1Q3F9J0 A0A182G5S9 A0A182IJS1 A0A023EN27 A0A1Q3F865 A0A1Q3G294 B0WKH0 A0A1Q3F7A6 A0A1Q3G251 A0A0L7LKH9 A0A0N0BJA3 A0A232EYV2 A0A0L7R1U6 E2A778 E9IML8 A0A088A648 A0A151I6R0 A0A151WP82 V9IE02 A0A151K112 A0A2A3EIZ8 A0A0J7KTJ1 T1D4M7 A0A151IV44 A0A195BAI1 K7J441 E2BHJ8 A0A1B6I671 A0A158NUV1 A0A1B6JPA2 A0A026W458 A0A1B6MG04 A0A1B6L1T3 A0A1B6G1E6 A0A0M4ENL2 A0A1B6MVC5 A0A067QSJ5 B3GC12 B3GC07 B3GBZ2 B3GC09 B3GBZ1 B4L9G8 A0A0T6AZA7 B4LB54 B4PEG7 E0VGF1 B3GBZ9 A0A1I8MEM7 Q29E07 T1PE16 A0A1I8P2T3 A0A0J9UIP9 B3M5K5 A0A1I8P2N4 A0A3B0JMK7 B4HDR1 B4H437 B4J0Y2
A0A2H1WW91 A0A194RH82 A0A182MEB2 A0A336M2W3 A0A336LZ35 A0A182P5E5 A0A182R6M1 A0A182SDB9 A0A182WDP9 A0A1Y9GZ48 T1EAE3 W5JW57 A0A182YAW9 A0A182FW20 A0A2M3Z716 A0A2M4AJ97 A0A182N251 A0A182QHB0 A0A182HW61 A0A182JP11 A0A182VP56 Q16RC6 A0A182X5T4 Q7PSP6 A0A182KKX1 A0A1L8DLB9 A0A2M4CHY8 A0A182UE94 U5ETQ0 A0A084W8C1 A0A1L8DL95 A0A1B0GIR7 A0A2M4CGY4 A0A0K8TNB1 T1E2Z8 A0A1Q3F9J0 A0A182G5S9 A0A182IJS1 A0A023EN27 A0A1Q3F865 A0A1Q3G294 B0WKH0 A0A1Q3F7A6 A0A1Q3G251 A0A0L7LKH9 A0A0N0BJA3 A0A232EYV2 A0A0L7R1U6 E2A778 E9IML8 A0A088A648 A0A151I6R0 A0A151WP82 V9IE02 A0A151K112 A0A2A3EIZ8 A0A0J7KTJ1 T1D4M7 A0A151IV44 A0A195BAI1 K7J441 E2BHJ8 A0A1B6I671 A0A158NUV1 A0A1B6JPA2 A0A026W458 A0A1B6MG04 A0A1B6L1T3 A0A1B6G1E6 A0A0M4ENL2 A0A1B6MVC5 A0A067QSJ5 B3GC12 B3GC07 B3GBZ2 B3GC09 B3GBZ1 B4L9G8 A0A0T6AZA7 B4LB54 B4PEG7 E0VGF1 B3GBZ9 A0A1I8MEM7 Q29E07 T1PE16 A0A1I8P2T3 A0A0J9UIP9 B3M5K5 A0A1I8P2N4 A0A3B0JMK7 B4HDR1 B4H437 B4J0Y2
PDB
1LVG
E-value=1.61508e-50,
Score=500
Ontologies
PATHWAY
GO
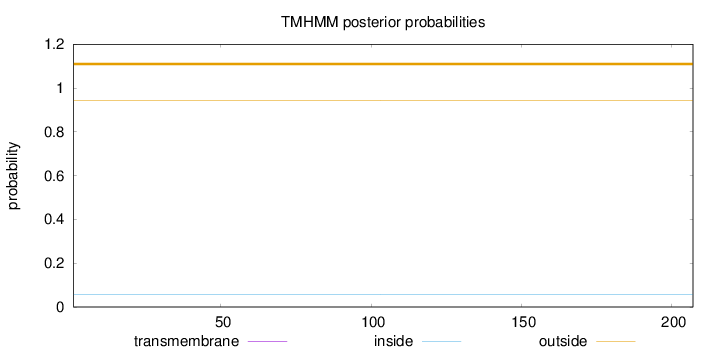
Topology
Length:
207
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00118
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.05846
outside
1 - 207
Population Genetic Test Statistics
Pi
216.65353
Theta
201.766601
Tajima's D
0.163619
CLR
0.296898
CSRT
0.418129093545323
Interpretation
Uncertain
