Gene
KWMTBOMO02669
Pre Gene Modal
BGIBMGA003448
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.879
Sequence
CDS
ATGTTAGGATTGGATTCGAAACTCGTCGGTCAAGCATTTCAACCGGAATTTGCTGAGCCATTGATGAATATAACGATACCAGTAGGACGAGATGCTACATTCAGATGCCTTGTACAGAACCTTGGAGGATACAGGGTAGGCTGGGTTAAAGCTGATACAAAGGCCATTCAAGCGATACATGTTCATGTCATAACCAACAACCATAGAGTCGCTGTGTCCCACAACGGGCCAACTGTTTGGAATCTACACATCCGTAGCGTTCAAGAAGAAGATAGAGGCCAATATATGTGTCAAATTAATACAGACCCAATGAAAAGCCAGATAGGTTACTTGGATGTAGTTGTATCACCCGACTTCATAGCGGAAGAAACGTCAGGTGATATTATGGTGCCAGAAGGAGGTACTGCTAGGGTTTCTTGTCGTGCTCGAGGTATGCCAGAGCCACGAGTCATGTGGCGGAGGGAAGATGGGGCTGATATCGTTTTGAGAGAGTCCAACGGCATCAAAACTAAAGTCGCTGTTTATGAAGAAGAAGTACTAACTTTAAACAAAATATCTCGATCAGACATGGGTGCATATTTGTGCATAGCGAGCAACGGCGTCCCGCCTTCTGTGAGCAAGAGAATCATCATAAAAGTTCATTTTCATCCCGTAATTCAAGTTCCAAATCAGCTGATCGGAGCACCTCTAGGCACCGATACTACCCTTGAATGCTACGTCGAGTCATCGCCGAAATCCATAAACTATTGGGTTCGAGATTCCAATGAGATGGTAATATCATCCAGCAAGTACGAAGTGATGAACACAGTTATAAGTTCGTTTGAGAGTCGCATGGCTTTGACTGTCCGAAGATTGACAGAAGAAGATATAGGCGGTTACCGGTGCGTTGCGAAGAATTCTCTGGGCGAAACCGATAGCCTCATTAGGATATATGAAATTCTCGGTCCAACAGTCAGAAACACGAGCCCGGTAACGAGGAGAGGAGACCTCAAGTACTCCACTCCCATAGAAGATCCCGACGAGCAGTTCGGTTCCGCCGAACGCTCTGACGACGAAGACGAGACCACTGACGTCACAGATCGGCTTCAGTACCAGACCAGGAATCGTACAGTACAACACACAACGAATTCGGCACACGAACAAAAACTAAGCAACAAAGTTAGGAAAATTAGCAATAAATTCGAAATTAAATCTGGATTTGAAACGAACAGCTGTTGCCGCGTACATTCCTATAATTTACATATCGCGATACTAGTTTTAAACTTTTATGCTTATAATTGA
Protein
MLGLDSKLVGQAFQPEFAEPLMNITIPVGRDATFRCLVQNLGGYRVGWVKADTKAIQAIHVHVITNNHRVAVSHNGPTVWNLHIRSVQEEDRGQYMCQINTDPMKSQIGYLDVVVSPDFIAEETSGDIMVPEGGTARVSCRARGMPEPRVMWRREDGADIVLRESNGIKTKVAVYEEEVLTLNKISRSDMGAYLCIASNGVPPSVSKRIIIKVHFHPVIQVPNQLIGAPLGTDTTLECYVESSPKSINYWVRDSNEMVISSSKYEVMNTVISSFESRMALTVRRLTEEDIGGYRCVAKNSLGETDSLIRIYEILGPTVRNTSPVTRRGDLKYSTPIEDPDEQFGSAERSDDEDETTDVTDRLQYQTRNRTVQHTTNSAHEQKLSNKVRKISNKFEIKSGFETNSCCRVHSYNLHIAILVLNFYAYN
Summary
Uniprot
A0A2A4JL61
A0A1E1W0X2
A0A2A4JJQ6
A0A212F5A5
A0A194RGV6
A0A194PP86
+ More
A0A139WP49 A0A182W618 A0A212FI49 A0A194PUH5 A0A182RVN5 A0A182FIH1 A0A2A4K4N0 A0A0M4ETJ3 A0A1I8Q817 B4MCZ3 A0A2S2Q2I9 B4JME1 B4I164 A0A1W4VT22 B3MW76 B3NSQ3 B4Q0J2 Q9W4R3 A0A3B0JW51 A0A0L0C9B6 A0A194RL15 A0A0R3P0U2 B4R463 A0A1J1I9R8 B5DNC9 A0A2H8TMT1 A0A0L7QR22 A0A158P1L0 A0A2A3EME1 A0A151WQF6 E2AQD4 A0A2S2Q9N1 A0A1B6MTQ2 D6W7P0 A0A1Y1L5Q2 A0A3L8DU39 A0A195D114 A0A1B6M242 A0A195BFJ2 A0A026X2K4 A0A2S2PES2 A0A139WPH8 A0A139WPG9 A0A2H8TUH6 A0A1Y1L4J8 A0A154PDF1 A0A212F5B1 Q5TQN9 K7IPY4 A0A194PPY2 Q17DU8 A0A2A4JLN5 A0A088AH34 A0A1S4GA51 A0A0C9RFT1 A0A084WFG5 A0A336KC27 F4X3S1 A0A158P039 E0VGC2 A0A3L8DWF9 A0A1I8MS04 A0A1W4V934 A0A151XJB3 A0A0L7QQE4 A0A1I8P3Y2 B4PZ42 A0A1J1IVL5 X2JCW3 A0A182R254 A0A182FJW8 A0A1D2N219 A0A3B0JBE6 N6UKG2 A0A195CQQ0 A0A0L0BTM0 U4U3R5 A0A1I8P3U6 A0A0N8NZT0 A0A1I8P3X7 A0A0A9X241 A0A1I8P3V2 A0A1W4UVS5 A0A1Y1L333 A0A0Q5TLC4 A0A1Y1KVW7 A0A182XYB1 A0A3Q0JKY0 A0A0R1EEC2
A0A139WP49 A0A182W618 A0A212FI49 A0A194PUH5 A0A182RVN5 A0A182FIH1 A0A2A4K4N0 A0A0M4ETJ3 A0A1I8Q817 B4MCZ3 A0A2S2Q2I9 B4JME1 B4I164 A0A1W4VT22 B3MW76 B3NSQ3 B4Q0J2 Q9W4R3 A0A3B0JW51 A0A0L0C9B6 A0A194RL15 A0A0R3P0U2 B4R463 A0A1J1I9R8 B5DNC9 A0A2H8TMT1 A0A0L7QR22 A0A158P1L0 A0A2A3EME1 A0A151WQF6 E2AQD4 A0A2S2Q9N1 A0A1B6MTQ2 D6W7P0 A0A1Y1L5Q2 A0A3L8DU39 A0A195D114 A0A1B6M242 A0A195BFJ2 A0A026X2K4 A0A2S2PES2 A0A139WPH8 A0A139WPG9 A0A2H8TUH6 A0A1Y1L4J8 A0A154PDF1 A0A212F5B1 Q5TQN9 K7IPY4 A0A194PPY2 Q17DU8 A0A2A4JLN5 A0A088AH34 A0A1S4GA51 A0A0C9RFT1 A0A084WFG5 A0A336KC27 F4X3S1 A0A158P039 E0VGC2 A0A3L8DWF9 A0A1I8MS04 A0A1W4V934 A0A151XJB3 A0A0L7QQE4 A0A1I8P3Y2 B4PZ42 A0A1J1IVL5 X2JCW3 A0A182R254 A0A182FJW8 A0A1D2N219 A0A3B0JBE6 N6UKG2 A0A195CQQ0 A0A0L0BTM0 U4U3R5 A0A1I8P3U6 A0A0N8NZT0 A0A1I8P3X7 A0A0A9X241 A0A1I8P3V2 A0A1W4UVS5 A0A1Y1L333 A0A0Q5TLC4 A0A1Y1KVW7 A0A182XYB1 A0A3Q0JKY0 A0A0R1EEC2
Pubmed
22118469
26354079
18362917
19820115
17994087
17550304
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26687361 26109357 26109356 30467080 30688651 26108605 15632085 21347285 20798317 28004739 30249741 24508170 12364791 20075255 17510324 24438588 21719571 20566863 25315136 27289101 23537049 25401762 26823975 25244985
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26687361 26109357 26109356 30467080 30688651 26108605 15632085 21347285 20798317 28004739 30249741 24508170 12364791 20075255 17510324 24438588 21719571 20566863 25315136 27289101 23537049 25401762 26823975 25244985
EMBL
NWSH01001156
PCG72324.1
GDQN01010475
JAT80579.1
PCG72325.1
AGBW02010215
+ More
OWR48921.1 KQ460207 KPJ16674.1 KQ459597 KPI94793.1 KQ971307 KYB29770.1 AGBW02008433 OWR53414.1 KPI94790.1 NWSH01000132 PCG79215.1 CP012528 ALC48222.1 CH940660 EDW58065.2 GGMS01002687 MBY71890.1 CH916371 EDV92466.1 CH480819 EDW53245.1 CH902625 EDV35221.1 CH954180 EDV45733.1 CM000162 EDX01276.1 AE014298 BT003528 AAF45880.2 AAO39532.1 AGB95064.1 OUUW01000011 SPP86305.1 JRES01000741 KNC28830.1 KPJ16671.1 CH379064 KRT06852.1 CM000366 EDX17005.1 CVRI01000044 CRK96482.1 EDY72936.1 GFXV01003679 MBW15484.1 KQ414784 KOC61077.1 ADTU01006465 ADTU01006466 ADTU01006467 ADTU01006468 ADTU01006469 KZ288210 PBC32973.1 KQ982828 KYQ50129.1 GL441723 EFN64348.1 GGMS01005057 MBY74260.1 GEBQ01000694 JAT39283.1 EFA11252.2 GEZM01068577 JAV66886.1 QOIP01000004 RLU23980.1 KQ976986 KYN06600.1 GEBQ01009995 JAT29982.1 KQ976490 KYM83350.1 KK107024 EZA62236.1 GGMR01015321 MBY27940.1 KYB29766.1 KYB29767.1 GFXV01006128 MBW17933.1 GEZM01068578 JAV66885.1 KQ434879 KZC09936.1 OWR48920.1 AAAB01008963 EAL39761.3 KPI94794.1 CH477291 EAT44564.1 PCG72322.1 GBYB01012002 JAG81769.1 ATLV01023347 ATLV01023348 KE525342 KFB48959.1 UFQS01000048 UFQT01000048 SSW98620.1 SSX19006.1 GL888624 EGI58868.1 ADTU01005306 ADTU01005307 ADTU01005308 ADTU01005309 ADTU01005310 ADTU01005311 DS235135 EEB12428.1 QOIP01000003 RLU24781.1 KQ982074 KYQ60502.1 KQ414787 KOC60799.1 EDX03103.2 CVRI01000061 CRL04144.1 AHN59990.1 LJIJ01000288 ODM99312.1 OUUW01000003 SPP77823.1 APGK01020360 KB740193 ENN81126.1 KQ977394 KYN03073.1 JRES01001480 KNC22574.1 KB631669 ERL85241.1 CH902622 KPU75180.1 GBHO01028807 GDHC01011086 JAG14797.1 JAQ07543.1 GEZM01072482 JAV65507.1 KQS30582.1 GEZM01072485 JAV65504.1 KRK07118.1
OWR48921.1 KQ460207 KPJ16674.1 KQ459597 KPI94793.1 KQ971307 KYB29770.1 AGBW02008433 OWR53414.1 KPI94790.1 NWSH01000132 PCG79215.1 CP012528 ALC48222.1 CH940660 EDW58065.2 GGMS01002687 MBY71890.1 CH916371 EDV92466.1 CH480819 EDW53245.1 CH902625 EDV35221.1 CH954180 EDV45733.1 CM000162 EDX01276.1 AE014298 BT003528 AAF45880.2 AAO39532.1 AGB95064.1 OUUW01000011 SPP86305.1 JRES01000741 KNC28830.1 KPJ16671.1 CH379064 KRT06852.1 CM000366 EDX17005.1 CVRI01000044 CRK96482.1 EDY72936.1 GFXV01003679 MBW15484.1 KQ414784 KOC61077.1 ADTU01006465 ADTU01006466 ADTU01006467 ADTU01006468 ADTU01006469 KZ288210 PBC32973.1 KQ982828 KYQ50129.1 GL441723 EFN64348.1 GGMS01005057 MBY74260.1 GEBQ01000694 JAT39283.1 EFA11252.2 GEZM01068577 JAV66886.1 QOIP01000004 RLU23980.1 KQ976986 KYN06600.1 GEBQ01009995 JAT29982.1 KQ976490 KYM83350.1 KK107024 EZA62236.1 GGMR01015321 MBY27940.1 KYB29766.1 KYB29767.1 GFXV01006128 MBW17933.1 GEZM01068578 JAV66885.1 KQ434879 KZC09936.1 OWR48920.1 AAAB01008963 EAL39761.3 KPI94794.1 CH477291 EAT44564.1 PCG72322.1 GBYB01012002 JAG81769.1 ATLV01023347 ATLV01023348 KE525342 KFB48959.1 UFQS01000048 UFQT01000048 SSW98620.1 SSX19006.1 GL888624 EGI58868.1 ADTU01005306 ADTU01005307 ADTU01005308 ADTU01005309 ADTU01005310 ADTU01005311 DS235135 EEB12428.1 QOIP01000003 RLU24781.1 KQ982074 KYQ60502.1 KQ414787 KOC60799.1 EDX03103.2 CVRI01000061 CRL04144.1 AHN59990.1 LJIJ01000288 ODM99312.1 OUUW01000003 SPP77823.1 APGK01020360 KB740193 ENN81126.1 KQ977394 KYN03073.1 JRES01001480 KNC22574.1 KB631669 ERL85241.1 CH902622 KPU75180.1 GBHO01028807 GDHC01011086 JAG14797.1 JAQ07543.1 GEZM01072482 JAV65507.1 KQS30582.1 GEZM01072485 JAV65504.1 KRK07118.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
UP000075920
+ More
UP000075900 UP000069272 UP000092553 UP000095300 UP000008792 UP000001070 UP000001292 UP000192221 UP000007801 UP000008711 UP000002282 UP000000803 UP000268350 UP000037069 UP000001819 UP000000304 UP000183832 UP000053825 UP000005205 UP000242457 UP000075809 UP000000311 UP000279307 UP000078542 UP000078540 UP000053097 UP000076502 UP000007062 UP000002358 UP000008820 UP000005203 UP000030765 UP000007755 UP000009046 UP000095301 UP000094527 UP000019118 UP000030742 UP000076408 UP000079169
UP000075900 UP000069272 UP000092553 UP000095300 UP000008792 UP000001070 UP000001292 UP000192221 UP000007801 UP000008711 UP000002282 UP000000803 UP000268350 UP000037069 UP000001819 UP000000304 UP000183832 UP000053825 UP000005205 UP000242457 UP000075809 UP000000311 UP000279307 UP000078542 UP000078540 UP000053097 UP000076502 UP000007062 UP000002358 UP000008820 UP000005203 UP000030765 UP000007755 UP000009046 UP000095301 UP000094527 UP000019118 UP000030742 UP000076408 UP000079169
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JL61
A0A1E1W0X2
A0A2A4JJQ6
A0A212F5A5
A0A194RGV6
A0A194PP86
+ More
A0A139WP49 A0A182W618 A0A212FI49 A0A194PUH5 A0A182RVN5 A0A182FIH1 A0A2A4K4N0 A0A0M4ETJ3 A0A1I8Q817 B4MCZ3 A0A2S2Q2I9 B4JME1 B4I164 A0A1W4VT22 B3MW76 B3NSQ3 B4Q0J2 Q9W4R3 A0A3B0JW51 A0A0L0C9B6 A0A194RL15 A0A0R3P0U2 B4R463 A0A1J1I9R8 B5DNC9 A0A2H8TMT1 A0A0L7QR22 A0A158P1L0 A0A2A3EME1 A0A151WQF6 E2AQD4 A0A2S2Q9N1 A0A1B6MTQ2 D6W7P0 A0A1Y1L5Q2 A0A3L8DU39 A0A195D114 A0A1B6M242 A0A195BFJ2 A0A026X2K4 A0A2S2PES2 A0A139WPH8 A0A139WPG9 A0A2H8TUH6 A0A1Y1L4J8 A0A154PDF1 A0A212F5B1 Q5TQN9 K7IPY4 A0A194PPY2 Q17DU8 A0A2A4JLN5 A0A088AH34 A0A1S4GA51 A0A0C9RFT1 A0A084WFG5 A0A336KC27 F4X3S1 A0A158P039 E0VGC2 A0A3L8DWF9 A0A1I8MS04 A0A1W4V934 A0A151XJB3 A0A0L7QQE4 A0A1I8P3Y2 B4PZ42 A0A1J1IVL5 X2JCW3 A0A182R254 A0A182FJW8 A0A1D2N219 A0A3B0JBE6 N6UKG2 A0A195CQQ0 A0A0L0BTM0 U4U3R5 A0A1I8P3U6 A0A0N8NZT0 A0A1I8P3X7 A0A0A9X241 A0A1I8P3V2 A0A1W4UVS5 A0A1Y1L333 A0A0Q5TLC4 A0A1Y1KVW7 A0A182XYB1 A0A3Q0JKY0 A0A0R1EEC2
A0A139WP49 A0A182W618 A0A212FI49 A0A194PUH5 A0A182RVN5 A0A182FIH1 A0A2A4K4N0 A0A0M4ETJ3 A0A1I8Q817 B4MCZ3 A0A2S2Q2I9 B4JME1 B4I164 A0A1W4VT22 B3MW76 B3NSQ3 B4Q0J2 Q9W4R3 A0A3B0JW51 A0A0L0C9B6 A0A194RL15 A0A0R3P0U2 B4R463 A0A1J1I9R8 B5DNC9 A0A2H8TMT1 A0A0L7QR22 A0A158P1L0 A0A2A3EME1 A0A151WQF6 E2AQD4 A0A2S2Q9N1 A0A1B6MTQ2 D6W7P0 A0A1Y1L5Q2 A0A3L8DU39 A0A195D114 A0A1B6M242 A0A195BFJ2 A0A026X2K4 A0A2S2PES2 A0A139WPH8 A0A139WPG9 A0A2H8TUH6 A0A1Y1L4J8 A0A154PDF1 A0A212F5B1 Q5TQN9 K7IPY4 A0A194PPY2 Q17DU8 A0A2A4JLN5 A0A088AH34 A0A1S4GA51 A0A0C9RFT1 A0A084WFG5 A0A336KC27 F4X3S1 A0A158P039 E0VGC2 A0A3L8DWF9 A0A1I8MS04 A0A1W4V934 A0A151XJB3 A0A0L7QQE4 A0A1I8P3Y2 B4PZ42 A0A1J1IVL5 X2JCW3 A0A182R254 A0A182FJW8 A0A1D2N219 A0A3B0JBE6 N6UKG2 A0A195CQQ0 A0A0L0BTM0 U4U3R5 A0A1I8P3U6 A0A0N8NZT0 A0A1I8P3X7 A0A0A9X241 A0A1I8P3V2 A0A1W4UVS5 A0A1Y1L333 A0A0Q5TLC4 A0A1Y1KVW7 A0A182XYB1 A0A3Q0JKY0 A0A0R1EEC2
PDB
6EFY
E-value=1.61102e-121,
Score=1116
Ontologies
GO
Topology
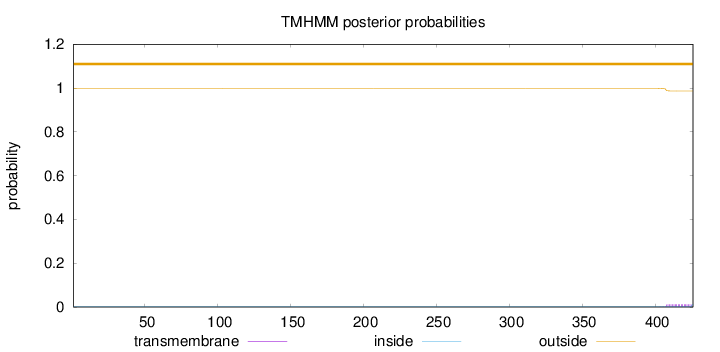
Length:
426
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22773
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00123
outside
1 - 426
Population Genetic Test Statistics
Pi
277.85667
Theta
199.761716
Tajima's D
1.542236
CLR
0.412054
CSRT
0.802509874506275
Interpretation
Uncertain
