Gene
KWMTBOMO02668
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.153
Sequence
CDS
ATGACCATTAACGCTGGCGTTCCACAAGGTTCGGTGCTCTCCCCCACGCTTTTCATCCTGTATATCAATGACATGCTGTCTATTGATGGCATGCATTGCTATGCGGATGACAGCACGGGGGATGCGCGATATATCGGCCATCAGAGTCTCTCTCGTGGCGTGGTGCAAGAAAGACGATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCCAAATGGGGTGAATTGAACTTGGTTCAATTCAACCCGTTAAAGACACAAGTTTGCGCGTTCACTGCGAAGAAGGACCCCTTTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTGCAACCTTCCGAGAGTATTGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCGAAGCGGTACTTCACGCCTGGACAAAGGCTTTTGCTTTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCGCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTCTTCCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCATTCTCACGGATCGTTTGGAGCCTCTGGGTCTGCGGAGGGACTTCGGTTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCGGCATCTCGTTTTTACCATCGCACCGCCCGCCACCGGAGTAGAGTTCATCCATACTACCTGGAGCCACTGCGGTCATCCACAGTGCGTTTCCAGAGGTCTTTTTTGCCACGTACCATCCGGCTATGGAATGAGCTCCCCTCCACGGTGTTTCCCGAGCGCTATGACATGTCCTTCTTCAAACGAGGCTTGTGGAGAGTATTAAACGGTAGGCAGCGGCTTGGCTCTGCCCCTGGCATTGCTGAAGTCCATGGGCGACGGTAA
Protein
MTINAGVPQGSVLSPTLFILYINDMLSIDGMHCYADDSTGDARYIGHQSLSRGVVQERRSKLVSEVENSLGRVSKWGELNLVQFNPLKTQVCAFTAKKDPFVMAPQFQGVSLQPSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYFTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPFDSIQRRAVRIVDNPILTDRLEPLGLRRDFGSLCILYRMFHGECSEELFEMIPASRFYHRTARHRSRVHPYYLEPLRSSTVRFQRSFLPRTIRLWNELPSTVFPERYDMSFFKRGLWRVLNGRQRLGSAPGIAEVHGRR
Summary
Uniprot
Q6UV17
Q5KTM5
A0A2H1VFZ3
A0A0P4VXU0
A0A3D5S1K1
A0A2H1WCG9
+ More
J9GT67 T2M6M7 A0A1Y1S3P2 A0A1E1XGQ7 A0A131Y4I8 A0A147BU27 A0A131Y4Q4 A0A147BJQ0 A0A1E1XNL1 A0A1E1XN74 A0A147BNQ9 A0A1E1XNW6 A0A1E1X2T1 A0A131XQR9 A0A147BJ69 A0A090XE26 A0A131Y3E3 A0A131XRE4 A0A131XT39 A0A0P4VVV8 A0A1E1X2P5 A0A1G4M3N1 A0A2G8KET5 A0A147BQ97 A0A2B4RTL1 A0A0P4VTV9 A0A2H9T3F5 A0A1G4M5I2 A0A147BJZ1 A0A147BNC9 A0A147BLD9 A0A1E1XGC7 A0A2H9T4Y5 A0A0K8RPQ6 A0A1E1XPS6 A0A2I0UC10 A0A2H9T4F1 A0A1E1X2R3 A0A1E1XM20 A0A147BBR8 A0A147BB29 A0A2H9T4Q5 A0A2H9T5P0 A0A147BMA3 A0A1E1XVU2 W4ZGQ7 A0A1E1XN53 W4ZIH4 A0A147BI04 A0A147BPB7 A0A131XV56 V5H8T0 A0A147BKN8 A0A147BMF5 A0A147BRL4 A0A2S6A1F0 K7EXB4 W4Z0S9 K7EZ61 A0A2H9T2R7 K7EW64 A0A147BJE1 K7F115 A0A2H9T2Z5 A0A1E1WW75 K7F016 A0A2I0U3N1 K7EYW0 A0A147BLD0 A0A210QEX7 K7EWB0 K7F0H6 K7F012 K7EWE8 A0A2B4RFN8 A0A3M0JFD2 A0A147BL51 A0A147BK71 A0A131XTA8 K7F0V3 W4Z5Z2 A0A210QQ08 K7EX25
J9GT67 T2M6M7 A0A1Y1S3P2 A0A1E1XGQ7 A0A131Y4I8 A0A147BU27 A0A131Y4Q4 A0A147BJQ0 A0A1E1XNL1 A0A1E1XN74 A0A147BNQ9 A0A1E1XNW6 A0A1E1X2T1 A0A131XQR9 A0A147BJ69 A0A090XE26 A0A131Y3E3 A0A131XRE4 A0A131XT39 A0A0P4VVV8 A0A1E1X2P5 A0A1G4M3N1 A0A2G8KET5 A0A147BQ97 A0A2B4RTL1 A0A0P4VTV9 A0A2H9T3F5 A0A1G4M5I2 A0A147BJZ1 A0A147BNC9 A0A147BLD9 A0A1E1XGC7 A0A2H9T4Y5 A0A0K8RPQ6 A0A1E1XPS6 A0A2I0UC10 A0A2H9T4F1 A0A1E1X2R3 A0A1E1XM20 A0A147BBR8 A0A147BB29 A0A2H9T4Q5 A0A2H9T5P0 A0A147BMA3 A0A1E1XVU2 W4ZGQ7 A0A1E1XN53 W4ZIH4 A0A147BI04 A0A147BPB7 A0A131XV56 V5H8T0 A0A147BKN8 A0A147BMF5 A0A147BRL4 A0A2S6A1F0 K7EXB4 W4Z0S9 K7EZ61 A0A2H9T2R7 K7EW64 A0A147BJE1 K7F115 A0A2H9T2Z5 A0A1E1WW75 K7F016 A0A2I0U3N1 K7EYW0 A0A147BLD0 A0A210QEX7 K7EWB0 K7F0H6 K7F012 K7EWE8 A0A2B4RFN8 A0A3M0JFD2 A0A147BL51 A0A147BK71 A0A131XTA8 K7F0V3 W4Z5Z2 A0A210QQ08 K7EX25
Pubmed
EMBL
AY359886
AAQ57129.1
AB126052
BAD86652.1
ODYU01002162
SOQ39302.1
+ More
GDRN01105059 JAI57791.1 DPOC01000269 HCX22453.1 ODYU01007423 SOQ50184.1 AMCI01002135 EJX03495.1 HAAD01001474 CDG67706.1 LWDP01000391 ORD92879.1 GFAC01000919 JAT98269.1 GEFM01002394 JAP73402.1 GEGO01001118 JAR94286.1 GEFM01002324 JAP73472.1 GEGO01004433 JAR90971.1 GFAA01002514 JAU00921.1 GFAA01002657 JAU00778.1 GEGO01003312 JAR92092.1 GFAA01002479 JAU00956.1 GFAC01005646 JAT93542.1 GEFM01006417 JAP69379.1 GEGO01004630 JAR90774.1 GBIH01002577 JAC92133.1 GEFM01002419 JAP73377.1 GEFM01005882 JAP69914.1 GEFM01006421 JAP69375.1 GDRN01111277 JAI56804.1 GFAC01005665 JAT93523.1 FLMD02000154 SCV66010.1 MRZV01000639 PIK46517.1 GEGO01002473 JAR92931.1 LSMT01000297 PFX20951.1 GDRN01111334 JAI56796.1 NSIT01000412 PJE77717.1 FLMD02000280 SCV66640.1 GEGO01004295 JAR91109.1 GEGO01003111 JAR92293.1 GEGO01003794 JAR91610.1 GFAC01000861 JAT98327.1 NSIT01000218 PJE78268.1 GADI01001229 JAA72579.1 GFAA01002385 JAU01050.1 KZ505886 PKU43598.1 NSIT01000271 PJE78081.1 GFAC01005670 JAT93518.1 GFAA01003161 JAU00274.1 GEGO01007187 JAR88217.1 GEGO01007438 JAR87966.1 NSIT01000239 PJE78188.1 NSIT01000164 PJE78524.1 GEGO01003518 JAR91886.1 GFAA01000066 JAU03369.1 AAGJ04096877 GFAA01002726 JAU00709.1 AAGJ04095885 GEGO01005479 JAR89925.1 GEGO01002794 JAR92610.1 GEFM01006250 JAP69546.1 GANP01010944 JAB73524.1 GEGO01004056 JAR91348.1 GEGO01003427 JAR91977.1 GEGO01002025 JAR93379.1 PSZC01000087 PPJ25136.1 AGCU01093320 AAGJ04088192 AGCU01081420 NSIT01000538 PJE77518.1 AGCU01055496 GEGO01004497 JAR90907.1 AGCU01164164 NSIT01000477 PJE77602.1 GFAC01007905 JAT91283.1 AGCU01148809 KZ506232 PKU40632.1 AGCU01046616 GEGO01004132 JAR91272.1 NEDP02004020 OWF47171.1 AGCU01168427 AGCU01096477 AGCU01135984 AGCU01003726 LSMT01000588 PFX15986.1 QRBI01000146 RMB99817.1 GEGO01003895 JAR91509.1 GEGO01004248 JAR91156.1 GEFM01005909 JAP69887.1 AGCU01111641 AAGJ04079835 NEDP02002425 OWF50811.1 AGCU01018596
GDRN01105059 JAI57791.1 DPOC01000269 HCX22453.1 ODYU01007423 SOQ50184.1 AMCI01002135 EJX03495.1 HAAD01001474 CDG67706.1 LWDP01000391 ORD92879.1 GFAC01000919 JAT98269.1 GEFM01002394 JAP73402.1 GEGO01001118 JAR94286.1 GEFM01002324 JAP73472.1 GEGO01004433 JAR90971.1 GFAA01002514 JAU00921.1 GFAA01002657 JAU00778.1 GEGO01003312 JAR92092.1 GFAA01002479 JAU00956.1 GFAC01005646 JAT93542.1 GEFM01006417 JAP69379.1 GEGO01004630 JAR90774.1 GBIH01002577 JAC92133.1 GEFM01002419 JAP73377.1 GEFM01005882 JAP69914.1 GEFM01006421 JAP69375.1 GDRN01111277 JAI56804.1 GFAC01005665 JAT93523.1 FLMD02000154 SCV66010.1 MRZV01000639 PIK46517.1 GEGO01002473 JAR92931.1 LSMT01000297 PFX20951.1 GDRN01111334 JAI56796.1 NSIT01000412 PJE77717.1 FLMD02000280 SCV66640.1 GEGO01004295 JAR91109.1 GEGO01003111 JAR92293.1 GEGO01003794 JAR91610.1 GFAC01000861 JAT98327.1 NSIT01000218 PJE78268.1 GADI01001229 JAA72579.1 GFAA01002385 JAU01050.1 KZ505886 PKU43598.1 NSIT01000271 PJE78081.1 GFAC01005670 JAT93518.1 GFAA01003161 JAU00274.1 GEGO01007187 JAR88217.1 GEGO01007438 JAR87966.1 NSIT01000239 PJE78188.1 NSIT01000164 PJE78524.1 GEGO01003518 JAR91886.1 GFAA01000066 JAU03369.1 AAGJ04096877 GFAA01002726 JAU00709.1 AAGJ04095885 GEGO01005479 JAR89925.1 GEGO01002794 JAR92610.1 GEFM01006250 JAP69546.1 GANP01010944 JAB73524.1 GEGO01004056 JAR91348.1 GEGO01003427 JAR91977.1 GEGO01002025 JAR93379.1 PSZC01000087 PPJ25136.1 AGCU01093320 AAGJ04088192 AGCU01081420 NSIT01000538 PJE77518.1 AGCU01055496 GEGO01004497 JAR90907.1 AGCU01164164 NSIT01000477 PJE77602.1 GFAC01007905 JAT91283.1 AGCU01148809 KZ506232 PKU40632.1 AGCU01046616 GEGO01004132 JAR91272.1 NEDP02004020 OWF47171.1 AGCU01168427 AGCU01096477 AGCU01135984 AGCU01003726 LSMT01000588 PFX15986.1 QRBI01000146 RMB99817.1 GEGO01003895 JAR91509.1 GEGO01004248 JAR91156.1 GEFM01005909 JAP69887.1 AGCU01111641 AAGJ04079835 NEDP02002425 OWF50811.1 AGCU01018596
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q6UV17
Q5KTM5
A0A2H1VFZ3
A0A0P4VXU0
A0A3D5S1K1
A0A2H1WCG9
+ More
J9GT67 T2M6M7 A0A1Y1S3P2 A0A1E1XGQ7 A0A131Y4I8 A0A147BU27 A0A131Y4Q4 A0A147BJQ0 A0A1E1XNL1 A0A1E1XN74 A0A147BNQ9 A0A1E1XNW6 A0A1E1X2T1 A0A131XQR9 A0A147BJ69 A0A090XE26 A0A131Y3E3 A0A131XRE4 A0A131XT39 A0A0P4VVV8 A0A1E1X2P5 A0A1G4M3N1 A0A2G8KET5 A0A147BQ97 A0A2B4RTL1 A0A0P4VTV9 A0A2H9T3F5 A0A1G4M5I2 A0A147BJZ1 A0A147BNC9 A0A147BLD9 A0A1E1XGC7 A0A2H9T4Y5 A0A0K8RPQ6 A0A1E1XPS6 A0A2I0UC10 A0A2H9T4F1 A0A1E1X2R3 A0A1E1XM20 A0A147BBR8 A0A147BB29 A0A2H9T4Q5 A0A2H9T5P0 A0A147BMA3 A0A1E1XVU2 W4ZGQ7 A0A1E1XN53 W4ZIH4 A0A147BI04 A0A147BPB7 A0A131XV56 V5H8T0 A0A147BKN8 A0A147BMF5 A0A147BRL4 A0A2S6A1F0 K7EXB4 W4Z0S9 K7EZ61 A0A2H9T2R7 K7EW64 A0A147BJE1 K7F115 A0A2H9T2Z5 A0A1E1WW75 K7F016 A0A2I0U3N1 K7EYW0 A0A147BLD0 A0A210QEX7 K7EWB0 K7F0H6 K7F012 K7EWE8 A0A2B4RFN8 A0A3M0JFD2 A0A147BL51 A0A147BK71 A0A131XTA8 K7F0V3 W4Z5Z2 A0A210QQ08 K7EX25
J9GT67 T2M6M7 A0A1Y1S3P2 A0A1E1XGQ7 A0A131Y4I8 A0A147BU27 A0A131Y4Q4 A0A147BJQ0 A0A1E1XNL1 A0A1E1XN74 A0A147BNQ9 A0A1E1XNW6 A0A1E1X2T1 A0A131XQR9 A0A147BJ69 A0A090XE26 A0A131Y3E3 A0A131XRE4 A0A131XT39 A0A0P4VVV8 A0A1E1X2P5 A0A1G4M3N1 A0A2G8KET5 A0A147BQ97 A0A2B4RTL1 A0A0P4VTV9 A0A2H9T3F5 A0A1G4M5I2 A0A147BJZ1 A0A147BNC9 A0A147BLD9 A0A1E1XGC7 A0A2H9T4Y5 A0A0K8RPQ6 A0A1E1XPS6 A0A2I0UC10 A0A2H9T4F1 A0A1E1X2R3 A0A1E1XM20 A0A147BBR8 A0A147BB29 A0A2H9T4Q5 A0A2H9T5P0 A0A147BMA3 A0A1E1XVU2 W4ZGQ7 A0A1E1XN53 W4ZIH4 A0A147BI04 A0A147BPB7 A0A131XV56 V5H8T0 A0A147BKN8 A0A147BMF5 A0A147BRL4 A0A2S6A1F0 K7EXB4 W4Z0S9 K7EZ61 A0A2H9T2R7 K7EW64 A0A147BJE1 K7F115 A0A2H9T2Z5 A0A1E1WW75 K7F016 A0A2I0U3N1 K7EYW0 A0A147BLD0 A0A210QEX7 K7EWB0 K7F0H6 K7F012 K7EWE8 A0A2B4RFN8 A0A3M0JFD2 A0A147BL51 A0A147BK71 A0A131XTA8 K7F0V3 W4Z5Z2 A0A210QQ08 K7EX25
Ontologies
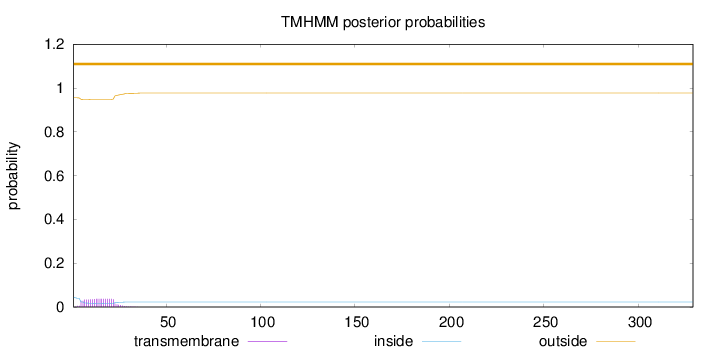
Topology
Length:
329
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.7135
Exp number, first 60 AAs:
0.71344
Total prob of N-in:
0.04468
outside
1 - 329
Population Genetic Test Statistics
Pi
145.499113
Theta
72.240282
Tajima's D
0.551275
CLR
0.619499
CSRT
0.534623268836558
Interpretation
Uncertain
