Gene
KWMTBOMO02665
Annotation
PREDICTED:_uncharacterized_protein_LOC106137189_isoform_X1_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.014 PlasmaMembrane Reliability : 1.53
Sequence
CDS
ATGCACCATCACACACTTCACATCGATGCTGGCGCTGGTGCCATTTCAGGAAATAATGCCAGCTCGGTGGCTCTTACCGTTTCTACAACTTCGACAGTCTTGCTCGGAACCGCAATTATACACGTCGCAGATTGTTGGGGCATTTATCAACCCGTACGTGCTATAATTGACAGTGGTGCCATGACGGGCTACATAACTCATGACTGTGCATTAAGGTTAGGACTAGGGCGAAGAAAGTGTGATTTCGATTCCATTGGTCTAGGTGCCAAGCCTATTGCAGATCTGGGGCTAACTACCTGTACTGTAAAACCGCGTCAGTGCAACAGCCCCATATTAACTACTGATGCAGTCATAGTTACGAAAATAGCCGGAAATCTACCAACTATGTCTATCTCAGATGATGTGATCCGTAAATTTCAAAATCTTTCACTTGCTGATCCAACTTTTGGTACATCGGGTCCCATCGACATGTTATTGGCCGGTGATCTATTTCCTCAAATTTATGATGGAGAAAAGATCATGCCACAAGATAAGAACATGCCAGTGGCTTTGCATTCCATTTTCGGCTACGTCATTGCAGGTCAAGTATCGATTTCGGGTCATAGAACTGTAGACACGGAAAATCATAATATCAAAAGCAATAATACAAAATCATTTGTTGCCCTTTCTATTAACGTGGAGCATGAAACGCAGTCTTTCTCGGAGACAGAAAGCATTTGCTTCAACGAACCACAGCATCCAGATAATATATCAGTAGGGACATTGTCTGATCGCGATCATAACCGTGATAAGAAAAGTAGATGTTTCGTAAACTACCCTTTGCAACCTTTTTATAAACTTGGTGTTTTATCTGGAGCAGAAGAACGTAAATATATTGATATAGAAAAGAAGTTGGAACGCGATCTGAATTTAAAATTCATTCATCATGCAAAACAATACATGAGTGGTTTAACGGCCTCCATCGGCTTAATGACTTCCAGTCAGAGGGTCCAGTTAAACCTATAA
Protein
MHHHTLHIDAGAGAISGNNASSVALTVSTTSTVLLGTAIIHVADCWGIYQPVRAIIDSGAMTGYITHDCALRLGLGRRKCDFDSIGLGAKPIADLGLTTCTVKPRQCNSPILTTDAVIVTKIAGNLPTMSISDDVIRKFQNLSLADPTFGTSGPIDMLLAGDLFPQIYDGEKIMPQDKNMPVALHSIFGYVIAGQVSISGHRTVDTENHNIKSNNTKSFVALSINVEHETQSFSETESICFNEPQHPDNISVGTLSDRDHNRDKKSRCFVNYPLQPFYKLGVLSGAEERKYIDIEKKLERDLNLKFIHHAKQYMSGLTASIGLMTSSQRVQLNL
Summary
Uniprot
A0A0A9Z001
A0A0K8SX96
A0A2S2NDE3
X1X5R1
X1X3D9
A0A2S2NZ32
+ More
J9JS75 A0A2S2QVB8 A0A2S2NIB3 A0A3Q0JHJ6 J9K0A1 A0A2S2NWE9 J9LSC7 A0A2S2P047 A0A2S2PRK4 A0A1U8N896 A0A2S2QTX5 A0A2S2QDN7 J9JKU0 A0A2S2R2J0 X1WVF9 A0A0A9Z275 J9K351 J9LNP3 J9LFA9 A0A2S2NNC8 V5GHM7 X1XU12 A0A0A9X461 A0A2S2QWS1 A0A146LUX6 A0A2S2NHU0 X1XTX9 X1WJS5 X1WUC7 A0A0A9XJ38 A0A2S2PC01 A0A0A9YZM3 A0A146L3Y2 A0A2S2NHP6 A0A2S2PNC2 A0A0A9YU87 A0A0A9YYJ5 V5G7F0 A0A0A9Y3D4 A0A0A9WW84 A0A2S2PC00 A0A0A9X8U7 A0A146L7F7 A0A0A9ZHU0 A0A0A9ZG52 A0A0A9WEJ9 A0A2S2PLW6 A0A0A9WH19 A0A146L5E8 A0A2S2PAF4 A0A2S2NPW7 A0A1B6GGX5 A0A0A9ZJJ6 A0A2S2NBR8 A0A0A9YCP2 X1XU46 A0A023F0B3 A0A023F0E2
J9JS75 A0A2S2QVB8 A0A2S2NIB3 A0A3Q0JHJ6 J9K0A1 A0A2S2NWE9 J9LSC7 A0A2S2P047 A0A2S2PRK4 A0A1U8N896 A0A2S2QTX5 A0A2S2QDN7 J9JKU0 A0A2S2R2J0 X1WVF9 A0A0A9Z275 J9K351 J9LNP3 J9LFA9 A0A2S2NNC8 V5GHM7 X1XU12 A0A0A9X461 A0A2S2QWS1 A0A146LUX6 A0A2S2NHU0 X1XTX9 X1WJS5 X1WUC7 A0A0A9XJ38 A0A2S2PC01 A0A0A9YZM3 A0A146L3Y2 A0A2S2NHP6 A0A2S2PNC2 A0A0A9YU87 A0A0A9YYJ5 V5G7F0 A0A0A9Y3D4 A0A0A9WW84 A0A2S2PC00 A0A0A9X8U7 A0A146L7F7 A0A0A9ZHU0 A0A0A9ZG52 A0A0A9WEJ9 A0A2S2PLW6 A0A0A9WH19 A0A146L5E8 A0A2S2PAF4 A0A2S2NPW7 A0A1B6GGX5 A0A0A9ZJJ6 A0A2S2NBR8 A0A0A9YCP2 X1XU46 A0A023F0B3 A0A023F0E2
EMBL
GBHO01006408
JAG37196.1
GBRD01007977
JAG57844.1
GGMR01002187
MBY14806.1
+ More
ABLF02006784 ABLF02043036 GGMR01009854 MBY22473.1 ABLF02004278 ABLF02004280 ABLF02065929 GGMS01012504 MBY81707.1 GGMR01003897 MBY16516.1 ABLF02067195 GGMR01008932 GGMR01010920 MBY21551.1 MBY23539.1 ABLF02034704 ABLF02034708 ABLF02034709 ABLF02044644 ABLF02055319 GGMR01010198 MBY22817.1 GGMR01019355 MBY31974.1 GGMS01011369 MBY80572.1 GGMS01006636 MBY75839.1 ABLF02021785 GGMS01014951 MBY84154.1 ABLF02026240 ABLF02055027 GBHO01007689 JAG35915.1 ABLF02025193 ABLF02040487 ABLF02059921 ABLF02067163 ABLF02003331 ABLF02003332 GGMR01005823 MBY18442.1 GALX01004947 JAB63519.1 ABLF02008553 ABLF02008554 GBHO01028830 JAG14774.1 GGMS01012379 MBY81582.1 GDHC01008459 JAQ10170.1 GGMR01003717 MBY16336.1 ABLF02042081 GBHO01024761 JAG18843.1 GGMR01014306 MBY26925.1 GBHO01007061 JAG36543.1 GDHC01016727 JAQ01902.1 GGMR01004029 MBY16648.1 GGMR01018294 MBY30913.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GBHO01012499 GBHO01012498 GBHO01012497 GBHO01007479 GBHO01007451 GBHO01007450 JAG31105.1 JAG31106.1 JAG31107.1 JAG36125.1 JAG36153.1 JAG36154.1 GALX01002411 JAB66055.1 GBHO01017458 JAG26146.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 GGMR01014351 MBY26970.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 GDHC01014268 JAQ04361.1 GBHO01000061 JAG43543.1 GBHO01000060 JAG43544.1 GBHO01037783 JAG05821.1 GGMR01017784 MBY30403.1 GBHO01037781 JAG05823.1 GDHC01016272 JAQ02357.1 GGMR01013824 MBY26443.1 GGMR01006575 MBY19194.1 GECZ01008071 JAS61698.1 GBHO01000059 JAG43545.1 GGMR01002024 MBY14643.1 GBHO01013680 JAG29924.1 ABLF02003339 ABLF02058001 GBBI01004463 JAC14249.1 GBBI01004318 JAC14394.1
ABLF02006784 ABLF02043036 GGMR01009854 MBY22473.1 ABLF02004278 ABLF02004280 ABLF02065929 GGMS01012504 MBY81707.1 GGMR01003897 MBY16516.1 ABLF02067195 GGMR01008932 GGMR01010920 MBY21551.1 MBY23539.1 ABLF02034704 ABLF02034708 ABLF02034709 ABLF02044644 ABLF02055319 GGMR01010198 MBY22817.1 GGMR01019355 MBY31974.1 GGMS01011369 MBY80572.1 GGMS01006636 MBY75839.1 ABLF02021785 GGMS01014951 MBY84154.1 ABLF02026240 ABLF02055027 GBHO01007689 JAG35915.1 ABLF02025193 ABLF02040487 ABLF02059921 ABLF02067163 ABLF02003331 ABLF02003332 GGMR01005823 MBY18442.1 GALX01004947 JAB63519.1 ABLF02008553 ABLF02008554 GBHO01028830 JAG14774.1 GGMS01012379 MBY81582.1 GDHC01008459 JAQ10170.1 GGMR01003717 MBY16336.1 ABLF02042081 GBHO01024761 JAG18843.1 GGMR01014306 MBY26925.1 GBHO01007061 JAG36543.1 GDHC01016727 JAQ01902.1 GGMR01004029 MBY16648.1 GGMR01018294 MBY30913.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GBHO01012499 GBHO01012498 GBHO01012497 GBHO01007479 GBHO01007451 GBHO01007450 JAG31105.1 JAG31106.1 JAG31107.1 JAG36125.1 JAG36153.1 JAG36154.1 GALX01002411 JAB66055.1 GBHO01017458 JAG26146.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 GGMR01014351 MBY26970.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 GDHC01014268 JAQ04361.1 GBHO01000061 JAG43543.1 GBHO01000060 JAG43544.1 GBHO01037783 JAG05821.1 GGMR01017784 MBY30403.1 GBHO01037781 JAG05823.1 GDHC01016272 JAQ02357.1 GGMR01013824 MBY26443.1 GGMR01006575 MBY19194.1 GECZ01008071 JAS61698.1 GBHO01000059 JAG43545.1 GGMR01002024 MBY14643.1 GBHO01013680 JAG29924.1 ABLF02003339 ABLF02058001 GBBI01004463 JAC14249.1 GBBI01004318 JAC14394.1
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR008737 Peptidase_asp_put
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR015416 Znf_H2C2_histone_UAS-bd
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR001969 Aspartic_peptidase_AS
IPR001878 Znf_CCHC
IPR007527 Znf_SWIM
IPR018289 MULE_transposase_dom
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR017907 Znf_RING_CS
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR008737 Peptidase_asp_put
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR015416 Znf_H2C2_histone_UAS-bd
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR001969 Aspartic_peptidase_AS
IPR001878 Znf_CCHC
IPR007527 Znf_SWIM
IPR018289 MULE_transposase_dom
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR017907 Znf_RING_CS
Gene 3D
ProteinModelPortal
A0A0A9Z001
A0A0K8SX96
A0A2S2NDE3
X1X5R1
X1X3D9
A0A2S2NZ32
+ More
J9JS75 A0A2S2QVB8 A0A2S2NIB3 A0A3Q0JHJ6 J9K0A1 A0A2S2NWE9 J9LSC7 A0A2S2P047 A0A2S2PRK4 A0A1U8N896 A0A2S2QTX5 A0A2S2QDN7 J9JKU0 A0A2S2R2J0 X1WVF9 A0A0A9Z275 J9K351 J9LNP3 J9LFA9 A0A2S2NNC8 V5GHM7 X1XU12 A0A0A9X461 A0A2S2QWS1 A0A146LUX6 A0A2S2NHU0 X1XTX9 X1WJS5 X1WUC7 A0A0A9XJ38 A0A2S2PC01 A0A0A9YZM3 A0A146L3Y2 A0A2S2NHP6 A0A2S2PNC2 A0A0A9YU87 A0A0A9YYJ5 V5G7F0 A0A0A9Y3D4 A0A0A9WW84 A0A2S2PC00 A0A0A9X8U7 A0A146L7F7 A0A0A9ZHU0 A0A0A9ZG52 A0A0A9WEJ9 A0A2S2PLW6 A0A0A9WH19 A0A146L5E8 A0A2S2PAF4 A0A2S2NPW7 A0A1B6GGX5 A0A0A9ZJJ6 A0A2S2NBR8 A0A0A9YCP2 X1XU46 A0A023F0B3 A0A023F0E2
J9JS75 A0A2S2QVB8 A0A2S2NIB3 A0A3Q0JHJ6 J9K0A1 A0A2S2NWE9 J9LSC7 A0A2S2P047 A0A2S2PRK4 A0A1U8N896 A0A2S2QTX5 A0A2S2QDN7 J9JKU0 A0A2S2R2J0 X1WVF9 A0A0A9Z275 J9K351 J9LNP3 J9LFA9 A0A2S2NNC8 V5GHM7 X1XU12 A0A0A9X461 A0A2S2QWS1 A0A146LUX6 A0A2S2NHU0 X1XTX9 X1WJS5 X1WUC7 A0A0A9XJ38 A0A2S2PC01 A0A0A9YZM3 A0A146L3Y2 A0A2S2NHP6 A0A2S2PNC2 A0A0A9YU87 A0A0A9YYJ5 V5G7F0 A0A0A9Y3D4 A0A0A9WW84 A0A2S2PC00 A0A0A9X8U7 A0A146L7F7 A0A0A9ZHU0 A0A0A9ZG52 A0A0A9WEJ9 A0A2S2PLW6 A0A0A9WH19 A0A146L5E8 A0A2S2PAF4 A0A2S2NPW7 A0A1B6GGX5 A0A0A9ZJJ6 A0A2S2NBR8 A0A0A9YCP2 X1XU46 A0A023F0B3 A0A023F0E2
Ontologies
KEGG
Topology
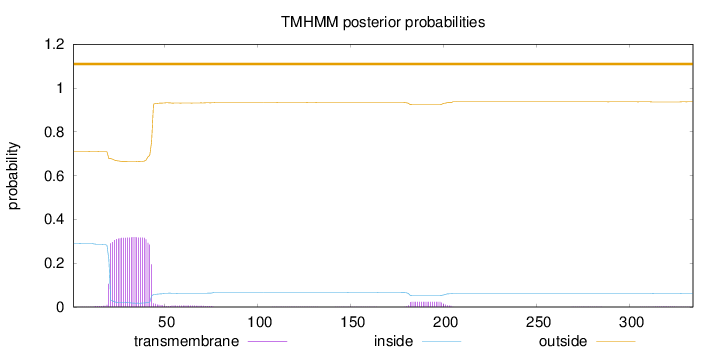
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.92118
Exp number, first 60 AAs:
7.31227
Total prob of N-in:
0.28974
outside
1 - 334
Population Genetic Test Statistics
Pi
218.874587
Theta
186.407352
Tajima's D
0.479529
CLR
111.203613
CSRT
0.504574771261437
Interpretation
Uncertain
