Gene
KWMTBOMO02664
Pre Gene Modal
BGIBMGA001503
Annotation
PREDICTED:_uncharacterized_protein_LOC105842132_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.675
Sequence
CDS
ATGGAGGCGGCGTGTCTCAAACGTATGGAGGATCTCAACCAAACCCATCGGAGAACCAGCAATAGTAGAATCCTTACCAAAAAACTCTGCGCCGGCAACAAACAATCTAGCACCAGAATAGGAATCTTACAAATTGAGAACATTTCATACCAGATTACAAACGACTACTCACAAATTGAAACGTTCTACCCATCTCTCAACTTTCTCTACGCCAATGTACGCAGCATTGTTAAGCCGGGAAAGTTTGATGAGCTCAAGTGCGTCCTGAAGTCTATCGATAAGGGTGTACACGTTGTTCTTCTCACCGAAACTTGGATCAAATCTGAAAATGATGCCATGAAACTACATTTGCCAAATTACACTCATCATTACAGCTATCGTAACGACATCAGAGGAGGCGGCGTGTCTATATACGTCCATAATAATATCAAACACAGCACGACTGAAGATAAATACAGCAATGGCAATAACTATCTTTGGGTGTATCTTGAGCAACATGGATTACACGTTGGAGTTGTATACAAGCCAGGAAGCACAAATACAGAAGATTTTCTTATCGATTATGAACAGCAACTACATGACAGAAATCGAACCATTATTTTCGGGGATTTCAACCTAGACCTATTACAAGAAGACAAACAAACTAAAATGTATAAACAAATGCTTTGCGAAAATGGATATAAAATATTGAATAAAGTAGAAGAAGAATACTGTACAAGAGAAACGTCGACAACTAGGACAATTTTGGATCATGTTACAACAAATTTAAAGAATAAAAACGTACACTTAGCCATTGTTGAATCTTGTATGTCGGATCATAAACAAATTTACGTCGCAATAAAAAAACAACAAGTAATTTCAAAACAAAGGAGTAAATACGAGGCCATAGACTATAGCAGCCTATATAAAAAATTTGATTCAAGCAAATTAGACAATAGTAACTATGAATACAAACATTTGGAAGATGTTATAAAACAGAATATTCGTGAAAACACTAAGACAAAATCGAAAATACTTAATTTGCCACAACAGGACTGGATAACAAAAAATATCACTGATGGAATAAATAGAAGAAATAAACTGTGGTACAAGCTGAAAAAGGACCCGAATAATGAAGACCTCAAAGCTAATTTCAAAACTGAAAGAAACCAGGCACAACCTTGGCCGATAAAATTCCTCAGAAATTTCATGGAAATCATACGCATACCTTACCAAATAACTCTGTGA
Protein
MEAACLKRMEDLNQTHRRTSNSRILTKKLCAGNKQSSTRIGILQIENISYQITNDYSQIETFYPSLNFLYANVRSIVKPGKFDELKCVLKSIDKGVHVVLLTETWIKSENDAMKLHLPNYTHHYSYRNDIRGGGVSIYVHNNIKHSTTEDKYSNGNNYLWVYLEQHGLHVGVVYKPGSTNTEDFLIDYEQQLHDRNRTIIFGDFNLDLLQEDKQTKMYKQMLCENGYKILNKVEEEYCTRETSTTRTILDHVTTNLKNKNVHLAIVESCMSDHKQIYVAIKKQQVISKQRSKYEAIDYSSLYKKFDSSKLDNSNYEYKHLEDVIKQNIRENTKTKSKILNLPQQDWITKNITDGINRRNKLWYKLKKDPNNEDLKANFKTERNQAQPWPIKFLRNFMEIIRIPYQITL
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9IW72
A0A0L7KTV2
A0A0L7LUD6
A0A0L7LHN9
A0A0L7LHZ6
A0A0L7KU39
+ More
A0A0L7KUP1 A0A0L7LL80 A0A0L7KU72 A0A2H1VEI6 A0A2A4JSM7 A0A0L7L5X3 A0A0L7LFP3 U5EHK5 A0A293N5E5 L7MAD0 L7MA61 L7MML1 L7MAV1 A0A293MNM5 A0A2R5LEU5 L7MGP0 A0A1B6KKL2 L7MDE9 A0A1J1IE11 W4Z536 T1FCX9 R7TYP7 R7TIU2 L7LST3 A0A147BLI8 A0A147BMP1 A0A3Q0JMB1 A0A0L7LQH0 R7VAZ6 A0A0L7KUG3 A0A147BMB8 A0A147BMP0 A0A293M0N9 A0A2S2NYK3 A0A293L920 A0A1B6HEU8 A0A147BCP6 A0A0L7LBE6 A0A147BLX9 A0A1J1IW52 A0A147BJM1 A0A147BWT2 A0A1W3JBA5 L7MMT2 A0A147BJU8 A0A131Y4P1 A0A0L7KQA2 A0A3P8N9X1 A0A3P8NF23 R7V082 A0A147BCF6 A0A293LDA4
A0A0L7KUP1 A0A0L7LL80 A0A0L7KU72 A0A2H1VEI6 A0A2A4JSM7 A0A0L7L5X3 A0A0L7LFP3 U5EHK5 A0A293N5E5 L7MAD0 L7MA61 L7MML1 L7MAV1 A0A293MNM5 A0A2R5LEU5 L7MGP0 A0A1B6KKL2 L7MDE9 A0A1J1IE11 W4Z536 T1FCX9 R7TYP7 R7TIU2 L7LST3 A0A147BLI8 A0A147BMP1 A0A3Q0JMB1 A0A0L7LQH0 R7VAZ6 A0A0L7KUG3 A0A147BMB8 A0A147BMP0 A0A293M0N9 A0A2S2NYK3 A0A293L920 A0A1B6HEU8 A0A147BCP6 A0A0L7LBE6 A0A147BLX9 A0A1J1IW52 A0A147BJM1 A0A147BWT2 A0A1W3JBA5 L7MMT2 A0A147BJU8 A0A131Y4P1 A0A0L7KQA2 A0A3P8N9X1 A0A3P8NF23 R7V082 A0A147BCF6 A0A293LDA4
EMBL
BABH01005955
BABH01005956
BABH01005957
JTDY01005746
KOB66672.1
JTDY01000106
+ More
KOB78836.1 JTDY01001032 KOB75078.1 KOB75077.1 JTDY01005835 KOB66566.1 JTDY01005656 KOB66776.1 JTDY01000656 KOB76313.1 JTDY01005628 KOB66807.1 ODYU01002150 SOQ39280.1 NWSH01000688 PCG74776.1 JTDY01002817 KOB70659.1 JTDY01001257 KOB74378.1 GANO01002979 JAB56892.1 GFWV01022122 MAA46850.1 GACK01003803 JAA61231.1 GACK01003878 JAA61156.1 GACK01000380 JAA64654.1 GACK01003889 JAA61145.1 GFWV01017766 MAA42494.1 GGLE01003890 MBY08016.1 GACK01002596 JAA62438.1 GEBQ01027994 JAT11983.1 GACK01003815 JAA61219.1 CVRI01000047 CRK98448.1 AAGJ04008561 AMQM01006321 KB097379 ESN97412.1 AMQN01026742 KB307264 ELT99048.1 AMQN01031095 AMQN01031096 KB310472 ELT91461.1 GACK01010154 JAA54880.1 GEGO01003737 JAR91667.1 GEGO01003346 JAR92058.1 JTDY01000313 KOB77743.1 AMQN01037559 KB293652 ELU15682.1 JTDY01005729 KOB66686.1 GEGO01003474 JAR91930.1 GEGO01003344 JAR92060.1 GFWV01009040 GFWV01009041 MAA33769.1 GGMR01009684 MBY22303.1 GFWV01000182 MAA24912.1 GECU01034449 JAS73257.1 GEGO01007172 JAR88232.1 JTDY01001811 KOB72812.1 GEGO01003604 JAR91800.1 CVRI01000059 CRL03366.1 GEGO01004417 JAR90987.1 GEGO01000196 JAR95208.1 GACK01000076 JAA64958.1 GEGO01004368 JAR91036.1 GEFM01002318 JAP73478.1 JTDY01007074 KOB65472.1 AMQN01005457 KB296106 ELU12238.1 GEGO01007379 JAR88025.1 GFWV01001118 MAA25848.1
KOB78836.1 JTDY01001032 KOB75078.1 KOB75077.1 JTDY01005835 KOB66566.1 JTDY01005656 KOB66776.1 JTDY01000656 KOB76313.1 JTDY01005628 KOB66807.1 ODYU01002150 SOQ39280.1 NWSH01000688 PCG74776.1 JTDY01002817 KOB70659.1 JTDY01001257 KOB74378.1 GANO01002979 JAB56892.1 GFWV01022122 MAA46850.1 GACK01003803 JAA61231.1 GACK01003878 JAA61156.1 GACK01000380 JAA64654.1 GACK01003889 JAA61145.1 GFWV01017766 MAA42494.1 GGLE01003890 MBY08016.1 GACK01002596 JAA62438.1 GEBQ01027994 JAT11983.1 GACK01003815 JAA61219.1 CVRI01000047 CRK98448.1 AAGJ04008561 AMQM01006321 KB097379 ESN97412.1 AMQN01026742 KB307264 ELT99048.1 AMQN01031095 AMQN01031096 KB310472 ELT91461.1 GACK01010154 JAA54880.1 GEGO01003737 JAR91667.1 GEGO01003346 JAR92058.1 JTDY01000313 KOB77743.1 AMQN01037559 KB293652 ELU15682.1 JTDY01005729 KOB66686.1 GEGO01003474 JAR91930.1 GEGO01003344 JAR92060.1 GFWV01009040 GFWV01009041 MAA33769.1 GGMR01009684 MBY22303.1 GFWV01000182 MAA24912.1 GECU01034449 JAS73257.1 GEGO01007172 JAR88232.1 JTDY01001811 KOB72812.1 GEGO01003604 JAR91800.1 CVRI01000059 CRL03366.1 GEGO01004417 JAR90987.1 GEGO01000196 JAR95208.1 GACK01000076 JAA64958.1 GEGO01004368 JAR91036.1 GEFM01002318 JAP73478.1 JTDY01007074 KOB65472.1 AMQN01005457 KB296106 ELU12238.1 GEGO01007379 JAR88025.1 GFWV01001118 MAA25848.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9IW72
A0A0L7KTV2
A0A0L7LUD6
A0A0L7LHN9
A0A0L7LHZ6
A0A0L7KU39
+ More
A0A0L7KUP1 A0A0L7LL80 A0A0L7KU72 A0A2H1VEI6 A0A2A4JSM7 A0A0L7L5X3 A0A0L7LFP3 U5EHK5 A0A293N5E5 L7MAD0 L7MA61 L7MML1 L7MAV1 A0A293MNM5 A0A2R5LEU5 L7MGP0 A0A1B6KKL2 L7MDE9 A0A1J1IE11 W4Z536 T1FCX9 R7TYP7 R7TIU2 L7LST3 A0A147BLI8 A0A147BMP1 A0A3Q0JMB1 A0A0L7LQH0 R7VAZ6 A0A0L7KUG3 A0A147BMB8 A0A147BMP0 A0A293M0N9 A0A2S2NYK3 A0A293L920 A0A1B6HEU8 A0A147BCP6 A0A0L7LBE6 A0A147BLX9 A0A1J1IW52 A0A147BJM1 A0A147BWT2 A0A1W3JBA5 L7MMT2 A0A147BJU8 A0A131Y4P1 A0A0L7KQA2 A0A3P8N9X1 A0A3P8NF23 R7V082 A0A147BCF6 A0A293LDA4
A0A0L7KUP1 A0A0L7LL80 A0A0L7KU72 A0A2H1VEI6 A0A2A4JSM7 A0A0L7L5X3 A0A0L7LFP3 U5EHK5 A0A293N5E5 L7MAD0 L7MA61 L7MML1 L7MAV1 A0A293MNM5 A0A2R5LEU5 L7MGP0 A0A1B6KKL2 L7MDE9 A0A1J1IE11 W4Z536 T1FCX9 R7TYP7 R7TIU2 L7LST3 A0A147BLI8 A0A147BMP1 A0A3Q0JMB1 A0A0L7LQH0 R7VAZ6 A0A0L7KUG3 A0A147BMB8 A0A147BMP0 A0A293M0N9 A0A2S2NYK3 A0A293L920 A0A1B6HEU8 A0A147BCP6 A0A0L7LBE6 A0A147BLX9 A0A1J1IW52 A0A147BJM1 A0A147BWT2 A0A1W3JBA5 L7MMT2 A0A147BJU8 A0A131Y4P1 A0A0L7KQA2 A0A3P8N9X1 A0A3P8NF23 R7V082 A0A147BCF6 A0A293LDA4
Ontologies
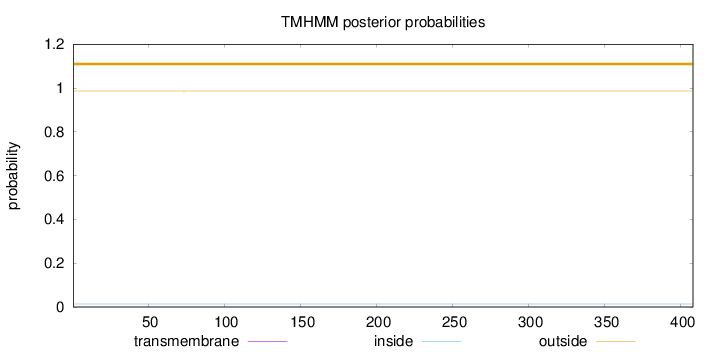
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00152
Exp number, first 60 AAs:
0.00031
Total prob of N-in:
0.01386
outside
1 - 408
Population Genetic Test Statistics
Pi
31.729443
Theta
163.410618
Tajima's D
0.713838
CLR
474.1681
CSRT
0.578221088945553
Interpretation
Uncertain
