Gene
KWMTBOMO02663
Pre Gene Modal
BGIBMGA003749
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.848 Nuclear Reliability : 1.516
Sequence
CDS
ATGAAAAGTCAAATGGGCTTCCTAGACGTAGTAATTCCTCCTGACTTTATACCTGAAGAAACGTCAGGGGACACCATGGTGCCCGAGGGAGGTACTGCAAGGGTGTCCTGCAAAGCACGAGGTGTTCCGCCTCCACGGGTGATGTGGAAAAGAGAGGACGGACAGGAAATCGTTGTGCGGGACCAGACTGGAGCAAAAACTAAAGGTAAATTTCATATATTGAACACTTCCAGAAATACAAAAAAATAA
Protein
MKSQMGFLDVVIPPDFIPEETSGDTMVPEGGTARVSCKARGVPPPRVMWKREDGQEIVVRDQTGAKTKGKFHILNTSRNTKK
Summary
Uniprot
H9J2L2
A0A2H1WU64
A0A2A4K4N0
A0A194PUH5
A0A212FI49
A0A2A4JJQ6
+ More
A0A2A4JL61 A0A0L7LKE7 A0A2H1W1K5 A0A1E1W0X2 H9J1R1 J9JRV5 A0A139WP49 E0VAF5 A0A067R3Q2 B4JME1 A0A1I8MHH7 A0A194RGV6 A0A194PP86 A0A212F5A5 A0A0L0C9B6 A0A1A9W1Y0 A0A1I8Q817 A0A182XYB1 A0A2P8YRY8 A0A2S2Q2I9 A0A2H8TMT1 A0A2P8XB09 A0A3L8DWF9 A0A0L7LS06 N6UIK1 A0A0L0BTM0 A0A1B0FFM0 A0A026WP64 A0A1I8P3Y2 B4H0U2 A0A1Y1L5Q2 A0A1W4VT22 A0A1I8MS04 A0A067R9I7 A0A1I8P3X7 A0A1I8P3V2 A0A0M4ETJ3 B3MW76 B3NSQ3 A0A1I8P3U6 Q9W4R3 A0A182MII5 A0A182RVN5 B4I164 A0A195F298 B4Q0J2 A0A182W618 A0A195E050 A0A3B0JW51 M9PGE1 B4MCZ3 A0A195CQQ0 A0A0R3P0U2 B4R463 B5DNC9 A0A1J1I9R8 A0A182SCE4 A0A182HBM4 B4NPW7 A0A2R7WK93 A0A182PAT7 A0A1B6M242 A0A1B6MTQ2 A0A182VVF9 A0A2S2Q9N1 A0A2P8ZE78 A0A182N0P2 A0A1B0D7N6 Q5TQN9 A0A1A9UHN8 A0A182WZ95 J9K816 A0A2H8TUH6 A0A2C9GQ43 A0A1S4GA51 A0A182ULY5 B0WSJ6 A0A336K2B0 A0A336KC27 A0A182TR57 E2B5A4 A0A1B0CRC5 A0A194RH74 A0A084WFG5 A0A182R254 K7IPY4 A0A182FIH1 A0A182FJW8 A0A1B6L2P2 A0A1J1IVL5 Q17DU8 E2AYH2 A0A2H1WEZ1
A0A2A4JL61 A0A0L7LKE7 A0A2H1W1K5 A0A1E1W0X2 H9J1R1 J9JRV5 A0A139WP49 E0VAF5 A0A067R3Q2 B4JME1 A0A1I8MHH7 A0A194RGV6 A0A194PP86 A0A212F5A5 A0A0L0C9B6 A0A1A9W1Y0 A0A1I8Q817 A0A182XYB1 A0A2P8YRY8 A0A2S2Q2I9 A0A2H8TMT1 A0A2P8XB09 A0A3L8DWF9 A0A0L7LS06 N6UIK1 A0A0L0BTM0 A0A1B0FFM0 A0A026WP64 A0A1I8P3Y2 B4H0U2 A0A1Y1L5Q2 A0A1W4VT22 A0A1I8MS04 A0A067R9I7 A0A1I8P3X7 A0A1I8P3V2 A0A0M4ETJ3 B3MW76 B3NSQ3 A0A1I8P3U6 Q9W4R3 A0A182MII5 A0A182RVN5 B4I164 A0A195F298 B4Q0J2 A0A182W618 A0A195E050 A0A3B0JW51 M9PGE1 B4MCZ3 A0A195CQQ0 A0A0R3P0U2 B4R463 B5DNC9 A0A1J1I9R8 A0A182SCE4 A0A182HBM4 B4NPW7 A0A2R7WK93 A0A182PAT7 A0A1B6M242 A0A1B6MTQ2 A0A182VVF9 A0A2S2Q9N1 A0A2P8ZE78 A0A182N0P2 A0A1B0D7N6 Q5TQN9 A0A1A9UHN8 A0A182WZ95 J9K816 A0A2H8TUH6 A0A2C9GQ43 A0A1S4GA51 A0A182ULY5 B0WSJ6 A0A336K2B0 A0A336KC27 A0A182TR57 E2B5A4 A0A1B0CRC5 A0A194RH74 A0A084WFG5 A0A182R254 K7IPY4 A0A182FIH1 A0A182FJW8 A0A1B6L2P2 A0A1J1IVL5 Q17DU8 E2AYH2 A0A2H1WEZ1
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
20566863 24845553 17994087 25315136 26108605 25244985 29403074 30249741 23537049 24508170 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26687361 26109357 26109356 30467080 30688651 17550304 15632085 26483478 12364791 20798317 24438588 20075255 17510324
20566863 24845553 17994087 25315136 26108605 25244985 29403074 30249741 23537049 24508170 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26687361 26109357 26109356 30467080 30688651 17550304 15632085 26483478 12364791 20798317 24438588 20075255 17510324
EMBL
BABH01008174
ODYU01011088
SOQ56605.1
NWSH01000132
PCG79215.1
KQ459597
+ More
KPI94790.1 AGBW02008433 OWR53414.1 NWSH01001156 PCG72325.1 PCG72324.1 JTDY01000798 KOB75839.1 ODYU01005716 SOQ46836.1 GDQN01010475 JAT80579.1 BABH01008165 ABLF02039546 ABLF02039548 ABLF02039558 ABLF02039559 ABLF02039563 ABLF02039572 ABLF02039575 KQ971307 KYB29770.1 DS235006 EEB10361.1 KK852725 KDR17668.1 CH916371 EDV92466.1 KQ460207 KPJ16674.1 KPI94793.1 AGBW02010215 OWR48921.1 JRES01000741 KNC28830.1 PYGN01000401 PSN46994.1 GGMS01002687 MBY71890.1 GFXV01003679 MBW15484.1 PYGN01005702 PSN29181.1 QOIP01000003 RLU24781.1 JTDY01000269 KOB77991.1 APGK01034033 APGK01034034 KB740901 ENN78472.1 JRES01001480 KNC22574.1 CCAG010008667 CCAG010008668 KK107139 EZA57847.1 CH479201 EDW30007.1 GEZM01068577 JAV66886.1 KK852607 KDR20347.1 CP012528 ALC48222.1 CH902625 EDV35221.1 CH954180 EDV45733.1 AE014298 BT003528 AAF45880.2 AAO39532.1 AGB95064.1 AXCM01001408 CH480819 EDW53245.1 KQ981856 KYN34506.1 CM000162 EDX01276.1 KQ979955 KYN18476.1 OUUW01000011 SPP86305.1 AGB95063.1 CH940660 EDW58065.2 KQ977394 KYN03073.1 CH379064 KRT06852.1 CM000366 EDX17005.1 EDY72936.1 CVRI01000044 CRK96482.1 JXUM01125309 JXUM01125310 JXUM01125311 KQ566931 KXJ69721.1 CH964291 EDW86192.2 KK854975 PTY20064.1 GEBQ01009995 JAT29982.1 GEBQ01000694 JAT39283.1 GGMS01005057 MBY74260.1 PYGN01000084 PSN54760.1 AJVK01012472 AJVK01012473 AAAB01008963 EAL39761.3 ABLF02029059 GFXV01006128 MBW17933.1 APCN01004867 DS232072 EDS33926.1 UFQS01000048 UFQT01000048 SSW98621.1 SSX19007.1 SSW98620.1 SSX19006.1 GL445761 EFN89131.1 AJWK01024595 AJWK01024596 AJWK01024597 KPJ16675.1 ATLV01023347 ATLV01023348 KE525342 KFB48959.1 GEBQ01022000 JAT17977.1 CVRI01000061 CRL04144.1 CH477291 EAT44564.1 GL443910 EFN61507.1 ODYU01008214 SOQ51621.1
KPI94790.1 AGBW02008433 OWR53414.1 NWSH01001156 PCG72325.1 PCG72324.1 JTDY01000798 KOB75839.1 ODYU01005716 SOQ46836.1 GDQN01010475 JAT80579.1 BABH01008165 ABLF02039546 ABLF02039548 ABLF02039558 ABLF02039559 ABLF02039563 ABLF02039572 ABLF02039575 KQ971307 KYB29770.1 DS235006 EEB10361.1 KK852725 KDR17668.1 CH916371 EDV92466.1 KQ460207 KPJ16674.1 KPI94793.1 AGBW02010215 OWR48921.1 JRES01000741 KNC28830.1 PYGN01000401 PSN46994.1 GGMS01002687 MBY71890.1 GFXV01003679 MBW15484.1 PYGN01005702 PSN29181.1 QOIP01000003 RLU24781.1 JTDY01000269 KOB77991.1 APGK01034033 APGK01034034 KB740901 ENN78472.1 JRES01001480 KNC22574.1 CCAG010008667 CCAG010008668 KK107139 EZA57847.1 CH479201 EDW30007.1 GEZM01068577 JAV66886.1 KK852607 KDR20347.1 CP012528 ALC48222.1 CH902625 EDV35221.1 CH954180 EDV45733.1 AE014298 BT003528 AAF45880.2 AAO39532.1 AGB95064.1 AXCM01001408 CH480819 EDW53245.1 KQ981856 KYN34506.1 CM000162 EDX01276.1 KQ979955 KYN18476.1 OUUW01000011 SPP86305.1 AGB95063.1 CH940660 EDW58065.2 KQ977394 KYN03073.1 CH379064 KRT06852.1 CM000366 EDX17005.1 EDY72936.1 CVRI01000044 CRK96482.1 JXUM01125309 JXUM01125310 JXUM01125311 KQ566931 KXJ69721.1 CH964291 EDW86192.2 KK854975 PTY20064.1 GEBQ01009995 JAT29982.1 GEBQ01000694 JAT39283.1 GGMS01005057 MBY74260.1 PYGN01000084 PSN54760.1 AJVK01012472 AJVK01012473 AAAB01008963 EAL39761.3 ABLF02029059 GFXV01006128 MBW17933.1 APCN01004867 DS232072 EDS33926.1 UFQS01000048 UFQT01000048 SSW98621.1 SSX19007.1 SSW98620.1 SSX19006.1 GL445761 EFN89131.1 AJWK01024595 AJWK01024596 AJWK01024597 KPJ16675.1 ATLV01023347 ATLV01023348 KE525342 KFB48959.1 GEBQ01022000 JAT17977.1 CVRI01000061 CRL04144.1 CH477291 EAT44564.1 GL443910 EFN61507.1 ODYU01008214 SOQ51621.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000007819
+ More
UP000007266 UP000009046 UP000027135 UP000001070 UP000095301 UP000053240 UP000037069 UP000091820 UP000095300 UP000076408 UP000245037 UP000279307 UP000019118 UP000092444 UP000053097 UP000008744 UP000192221 UP000092553 UP000007801 UP000008711 UP000000803 UP000075883 UP000075900 UP000001292 UP000078541 UP000002282 UP000075920 UP000078492 UP000268350 UP000008792 UP000078542 UP000001819 UP000000304 UP000183832 UP000075901 UP000069940 UP000249989 UP000007798 UP000075885 UP000075884 UP000092462 UP000007062 UP000078200 UP000076407 UP000075840 UP000075903 UP000002320 UP000075902 UP000008237 UP000092461 UP000030765 UP000002358 UP000069272 UP000008820 UP000000311
UP000007266 UP000009046 UP000027135 UP000001070 UP000095301 UP000053240 UP000037069 UP000091820 UP000095300 UP000076408 UP000245037 UP000279307 UP000019118 UP000092444 UP000053097 UP000008744 UP000192221 UP000092553 UP000007801 UP000008711 UP000000803 UP000075883 UP000075900 UP000001292 UP000078541 UP000002282 UP000075920 UP000078492 UP000268350 UP000008792 UP000078542 UP000001819 UP000000304 UP000183832 UP000075901 UP000069940 UP000249989 UP000007798 UP000075885 UP000075884 UP000092462 UP000007062 UP000078200 UP000076407 UP000075840 UP000075903 UP000002320 UP000075902 UP000008237 UP000092461 UP000030765 UP000002358 UP000069272 UP000008820 UP000000311
Interpro
Gene 3D
ProteinModelPortal
H9J2L2
A0A2H1WU64
A0A2A4K4N0
A0A194PUH5
A0A212FI49
A0A2A4JJQ6
+ More
A0A2A4JL61 A0A0L7LKE7 A0A2H1W1K5 A0A1E1W0X2 H9J1R1 J9JRV5 A0A139WP49 E0VAF5 A0A067R3Q2 B4JME1 A0A1I8MHH7 A0A194RGV6 A0A194PP86 A0A212F5A5 A0A0L0C9B6 A0A1A9W1Y0 A0A1I8Q817 A0A182XYB1 A0A2P8YRY8 A0A2S2Q2I9 A0A2H8TMT1 A0A2P8XB09 A0A3L8DWF9 A0A0L7LS06 N6UIK1 A0A0L0BTM0 A0A1B0FFM0 A0A026WP64 A0A1I8P3Y2 B4H0U2 A0A1Y1L5Q2 A0A1W4VT22 A0A1I8MS04 A0A067R9I7 A0A1I8P3X7 A0A1I8P3V2 A0A0M4ETJ3 B3MW76 B3NSQ3 A0A1I8P3U6 Q9W4R3 A0A182MII5 A0A182RVN5 B4I164 A0A195F298 B4Q0J2 A0A182W618 A0A195E050 A0A3B0JW51 M9PGE1 B4MCZ3 A0A195CQQ0 A0A0R3P0U2 B4R463 B5DNC9 A0A1J1I9R8 A0A182SCE4 A0A182HBM4 B4NPW7 A0A2R7WK93 A0A182PAT7 A0A1B6M242 A0A1B6MTQ2 A0A182VVF9 A0A2S2Q9N1 A0A2P8ZE78 A0A182N0P2 A0A1B0D7N6 Q5TQN9 A0A1A9UHN8 A0A182WZ95 J9K816 A0A2H8TUH6 A0A2C9GQ43 A0A1S4GA51 A0A182ULY5 B0WSJ6 A0A336K2B0 A0A336KC27 A0A182TR57 E2B5A4 A0A1B0CRC5 A0A194RH74 A0A084WFG5 A0A182R254 K7IPY4 A0A182FIH1 A0A182FJW8 A0A1B6L2P2 A0A1J1IVL5 Q17DU8 E2AYH2 A0A2H1WEZ1
A0A2A4JL61 A0A0L7LKE7 A0A2H1W1K5 A0A1E1W0X2 H9J1R1 J9JRV5 A0A139WP49 E0VAF5 A0A067R3Q2 B4JME1 A0A1I8MHH7 A0A194RGV6 A0A194PP86 A0A212F5A5 A0A0L0C9B6 A0A1A9W1Y0 A0A1I8Q817 A0A182XYB1 A0A2P8YRY8 A0A2S2Q2I9 A0A2H8TMT1 A0A2P8XB09 A0A3L8DWF9 A0A0L7LS06 N6UIK1 A0A0L0BTM0 A0A1B0FFM0 A0A026WP64 A0A1I8P3Y2 B4H0U2 A0A1Y1L5Q2 A0A1W4VT22 A0A1I8MS04 A0A067R9I7 A0A1I8P3X7 A0A1I8P3V2 A0A0M4ETJ3 B3MW76 B3NSQ3 A0A1I8P3U6 Q9W4R3 A0A182MII5 A0A182RVN5 B4I164 A0A195F298 B4Q0J2 A0A182W618 A0A195E050 A0A3B0JW51 M9PGE1 B4MCZ3 A0A195CQQ0 A0A0R3P0U2 B4R463 B5DNC9 A0A1J1I9R8 A0A182SCE4 A0A182HBM4 B4NPW7 A0A2R7WK93 A0A182PAT7 A0A1B6M242 A0A1B6MTQ2 A0A182VVF9 A0A2S2Q9N1 A0A2P8ZE78 A0A182N0P2 A0A1B0D7N6 Q5TQN9 A0A1A9UHN8 A0A182WZ95 J9K816 A0A2H8TUH6 A0A2C9GQ43 A0A1S4GA51 A0A182ULY5 B0WSJ6 A0A336K2B0 A0A336KC27 A0A182TR57 E2B5A4 A0A1B0CRC5 A0A194RH74 A0A084WFG5 A0A182R254 K7IPY4 A0A182FIH1 A0A182FJW8 A0A1B6L2P2 A0A1J1IVL5 Q17DU8 E2AYH2 A0A2H1WEZ1
PDB
6EFY
E-value=4.58108e-22,
Score=251
Ontologies
GO
Topology
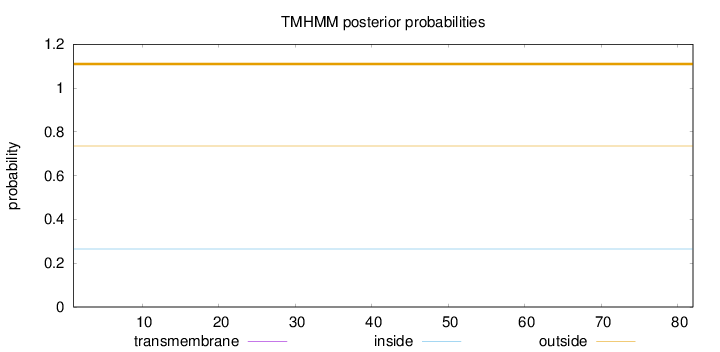
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.26477
outside
1 - 82
Population Genetic Test Statistics
Pi
278.633202
Theta
190.816388
Tajima's D
1.326215
CLR
0.156589
CSRT
0.742012899355032
Interpretation
Uncertain
