Gene
KWMTBOMO02662
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.785
Sequence
CDS
ATGTTCCGCGCTTCATTCAGGTGTTGCCTCGATCTCACCTCCAACATCCTACCTCTCCTAATCCTACTCTCCGAAAACTTAAACATCATGAAGACGACAAAAAAAAAGAAAAGGGTTTCACAGGCGGTCCCCCATTCCACATTTAATGTGGATTTCAGCAATGTCAGGGGCCTACATAGCAACCTTGACGCCGTACACCACCACCTTGAGACGGCGCAGCCTGCCCTGCTTTTTTTAACGGAGACGCAGATATCTGCTCCGGATGATACCTCATACCTTGAATACCCCGGCTACGTATTGGAGCACAACTTCCTGCGTAAAGCCGGGGTGTACGTATTCGTCCGGGCTGATGTCTGTTGTCGCCGTCTTCGGGGCCTCGAACAACGGGACCTGTCCCTCTTGTGGCTGCGCGTAGATCACGGGGGCCGTAGCCGAATCTACGCGTGCCTGTACAGGTCCCACAGCAGTGATGCAGGTTCAGCTCTGTTTGAGCATGTGCAAGAGGGGACTAACCGCGTGCTTGAGCAGTACCCATCTGCGGAGGTGGTGGTTCTTGGAGACTTTAACGCTCACCACCAAGAGTGGTTGGGATCCAGAACCACTGACCTCCCGGGTCGGACTGCCTACGATTTCGCCTTGGCCTACGGCTTCTCCCAGCTGGTGACACAGCCCACCCGTATCCCAGATATCGAGGGGCACGAGCCTTCTTTGTTGGACCTTCTGCTGACCACAGATCCGGTCGGATACAGTGTGGTGGTCGACGCTCCGCTTGGATCGTCTGATCACTGCCTTATTCGTGCTGCCGTACCACTCTCTCGTCCTGATCGTCGAACGACGACCAGGTATCGAAGAGTTTGGCGGTATATGTCAGCAGACTGGGATGGATTGCGTGAATTTTACGCATCCTACCCATGGGGGCGGTTCTGCTTTTCCTCTGCTGATCCTGACGTCTGTGCGGACCGTCTTAAAGACGTGGTGCTCCAGGGGATGGAATTGTTTATTCCCTCCTCTGAAGTGCCCGTTGGGGGTCGCAGCAGACCCTGGTATAACAATGCCAGCAGGGATGCTGCACACTTCAAGCGGTCCGCATACGTGGCATGGGATAATGCCAGGAGACGTCGGGATCCTAACATCTCAGGGGAAAGGCGGAAATATAACGCCGCTTCCAGGTCCTACAAGAAGGTAATTGCCAAGGCGAAGTCGGAGCACGTCGCTAGAGTTGGCGAGCGATTGAAGAGCTATCCCTCCGGGAGCCGTGCTTTTTGGTCGCTCGCCAAAGCTGCAGAAGGAAACTTCTGCAGGTCTAGTCTCCCACCACTGCGCAAGTCCGATGACAATCTGGCCCACAGCGCGAAAGAGAAGGCTGACCTTCTGGTCAAACTCTTCGCCTCGAACTCGACTGTGGACGACGGGGGTGCCACACCACCGAACATCCCCCGGTGTGATAGCTCCCTGCCGGAAATCTGCTTCACACAGTGTGCAGTCAGGTGGGAGCTCAGACTCCTGGACGTCCATAAGTCGAGTGGGCCAGACGGCATCCCCGCAGTGGTTTTGAAAACGTGCGCCCCTGAGCTGACACCTGCGCTAACGCGTTTGTATCGCCTCTCTTATTGCACTAACAGGGTTCCGTCTTCATGGAAGACCGCCCACGTCCACCCTATCCCCAAGAAGGGTGACCGGTCGGACCCATCGAGCTACAGGCCTATCGCGATAACTTCCTTGCTTTCCAAGGTGATGGAGCGAATTATAAATACACAACTCCTGAAGTATCTTGAAGATCGCCAGCTGATCAGTGACCGACAGTACGGTTTCCGTCACGGTCGCTCAGCTGGCGATCTTCTTGTATACCTTACTCACAGGTGGGCCTTGGAGAGCAAGGGCGAGGCTCTTGCTGTGAGCCTTGATATCGCGAAGGCCTTCGACAGGGTCTGGCATAGGGCACTTCTATCGAAGTTACCATCTTACGGAATCCCCGAGGGTCTCTGCAAGTGGATCGCTAGCTTTTTGGATGGGCGGAGCATCACGGTCGTTGTAGACGGTGACTGTTCTGATACCATGACCATTAACGCTGGCGTTCCACAAGGTTCGGTGCTCTCCCCCACGCTTTTCATCCTGTATATCAATGACATGCTGTCTATTGATGGCATGCATTGCTATGCAGATGATAGCACGGGGGATGCGCGATATATCGGCCATCAGAGTCTCTCTCGGAGCGTGGTGCAAGAGAGACGATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCCAAATGGGGTGAATCGAACTTAGTTCAATTCAACCCGTTGAAGACACAGGTTTGCGCGTTCACTGCGAAGAAGGACCCCTTTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTGCAACCTTCCGAGAGTATCGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCGAAGCGGTACTTCACGCCTGGACAAAGACTTTTGCTTTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTCTTCCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCATTCTCTCGGATCGTTTGGAGCCTCTGGGTCTGCGGAGGGACTTCGGTTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCAGCATCTCGTTTTTACCATCGCACCGCCCGCCACCGGAGTAGAGTTCATCCATACTACCTGGAGCCACTGCGGTCATCCACAGTGCGTTTCCAGAGATCTTTTTTGCCACGTACCATCCGGCTATGGAATGAGCTCCCCTCCACGGTATTTCCCGAGCGCTATGACATGTCCTTCTTCAAACGAGGCTTATGGAGAGTATTAAGCGGTAGGCAGCGGCTTGGCTCTGCCCCTGGCATTGCTGAAGTCCATGGGCGACGGTAA
Protein
MFRASFRCCLDLTSNILPLLILLSENLNIMKTTKKKKRVSQAVPHSTFNVDFSNVRGLHSNLDAVHHHLETAQPALLFLTETQISAPDDTSYLEYPGYVLEHNFLRKAGVYVFVRADVCCRRLRGLEQRDLSLLWLRVDHGGRSRIYACLYRSHSSDAGSALFEHVQEGTNRVLEQYPSAEVVVLGDFNAHHQEWLGSRTTDLPGRTAYDFALAYGFSQLVTQPTRIPDIEGHEPSLLDLLLTTDPVGYSVVVDAPLGSSDHCLIRAAVPLSRPDRRTTTRYRRVWRYMSADWDGLREFYASYPWGRFCFSSADPDVCADRLKDVVLQGMELFIPSSEVPVGGRSRPWYNNASRDAAHFKRSAYVAWDNARRRRDPNISGERRKYNAASRSYKKVIAKAKSEHVARVGERLKSYPSGSRAFWSLAKAAEGNFCRSSLPPLRKSDDNLAHSAKEKADLLVKLFASNSTVDDGGATPPNIPRCDSSLPEICFTQCAVRWELRLLDVHKSSGPDGIPAVVLKTCAPELTPALTRLYRLSYCTNRVPSSWKTAHVHPIPKKGDRSDPSSYRPIAITSLLSKVMERIINTQLLKYLEDRQLISDRQYGFRHGRSAGDLLVYLTHRWALESKGEALAVSLDIAKAFDRVWHRALLSKLPSYGIPEGLCKWIASFLDGRSITVVVDGDCSDTMTINAGVPQGSVLSPTLFILYINDMLSIDGMHCYADDSTGDARYIGHQSLSRSVVQERRSKLVSEVENSLGRVSKWGESNLVQFNPLKTQVCAFTAKKDPFVMAPQFQGVSLQPSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYFTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPFDSIQRRAVRIVDNPILSDRLEPLGLRRDFGSLCILYRMFHGECSEELFEMIPASRFYHRTARHRSRVHPYYLEPLRSSTVRFQRSFLPRTIRLWNELPSTVFPERYDMSFFKRGLWRVLSGRQRLGSAPGIAEVHGRR
Summary
Uniprot
EMBL
AY359886
AAQ57129.1
DPOC01000269
HCX22453.1
HAAD01000593
CDG66825.1
+ More
GDRN01111277 JAI56804.1 GDRN01111278 JAI56803.1 NSIT01000234 PJE78200.1 GDRN01111326 JAI56798.1 AAGJ04060226 AAGJ04103823 NSIT01000225 PJE78229.1 AAGJ04112043 NSIT01000271 PJE78081.1 AH011247 AAL40415.1 NSIT01000164 PJE78524.1
GDRN01111277 JAI56804.1 GDRN01111278 JAI56803.1 NSIT01000234 PJE78200.1 GDRN01111326 JAI56798.1 AAGJ04060226 AAGJ04103823 NSIT01000225 PJE78229.1 AAGJ04112043 NSIT01000271 PJE78081.1 AH011247 AAL40415.1 NSIT01000164 PJE78524.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=0.00034485,
Score=108
Ontologies
PANTHER
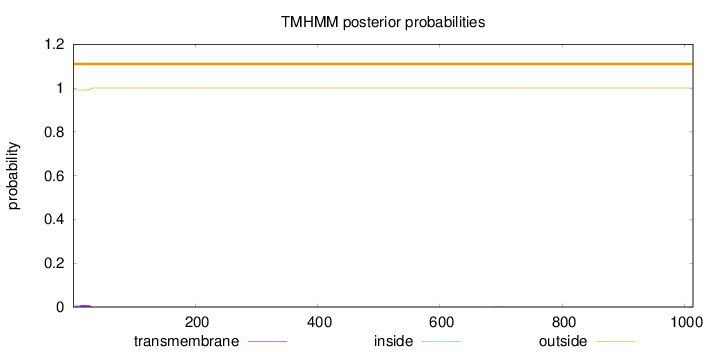
Topology
Length:
1014
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17353
Exp number, first 60 AAs:
0.17044
Total prob of N-in:
0.00896
outside
1 - 1014
Population Genetic Test Statistics
Pi
15.595916
Theta
105.940984
Tajima's D
0.52684
CLR
0.363874
CSRT
0.520073996300185
Interpretation
Uncertain
