Gene
KWMTBOMO02661
Pre Gene Modal
BGIBMGA003750
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.215
Sequence
CDS
ATGTCTGTACGTCAGAAGTCCATGTTCGAAGCCGAAATGAGAATGACGATAAAAAATATACGACGCGAAGACTTGGGAAGCTACATTTGTGTCGCGAAAAACTCGCTCGGCGATGTCGAGAGCAAAATAAGGTTATATGAAATACCAGGGAATGATAGACATTCGTACCATTACCCTGAGGTTCGGGTACCGACCGAGGACGACTACGGCACCGACAGCTATGACGATATGGAAGTCGAGGACAACGACAGCACTAAAACGAATAGGGTACCAGACCACGCCAATAAATGGTACGCTAATGACGGAAAGGTAATCGTTTCACAGTCCAACGATGCGTTCAAACCAACACCGTCATTCCTGACAATCATAACGATCTTCCTACTTCACAATAATCCAGTATAA
Protein
MSVRQKSMFEAEMRMTIKNIRREDLGSYICVAKNSLGDVESKIRLYEIPGNDRHSYHYPEVRVPTEDDYGTDSYDDMEVEDNDSTKTNRVPDHANKWYANDGKVIVSQSNDAFKPTPSFLTIITIFLLHNNPV
Summary
Uniprot
EMBL
BABH01008175
KQ459597
KPI94790.1
NWSH01000132
PCG79215.1
KQ460207
+ More
KPJ16671.1 ODYU01012414 SOQ58707.1 AGBW02008433 OWR53414.1 JTDY01001086 KOB74922.1 JXUM01054226 KQ561811 KXJ77453.1 AJWK01024595 AJWK01024596 AJWK01024597 DS233679 EDS31102.1 JXUM01111613 JXUM01111614 JXUM01111615 JXUM01111616 KQ565521 KXJ70905.1 KPI94794.1 GL441723 EFN64348.1 KK115496 KFM65269.1 GDQN01010475 JAT80579.1
KPJ16671.1 ODYU01012414 SOQ58707.1 AGBW02008433 OWR53414.1 JTDY01001086 KOB74922.1 JXUM01054226 KQ561811 KXJ77453.1 AJWK01024595 AJWK01024596 AJWK01024597 DS233679 EDS31102.1 JXUM01111613 JXUM01111614 JXUM01111615 JXUM01111616 KQ565521 KXJ70905.1 KPI94794.1 GL441723 EFN64348.1 KK115496 KFM65269.1 GDQN01010475 JAT80579.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
PDB
6EFY
E-value=4.65634e-10,
Score=148
Ontologies
GO
Topology
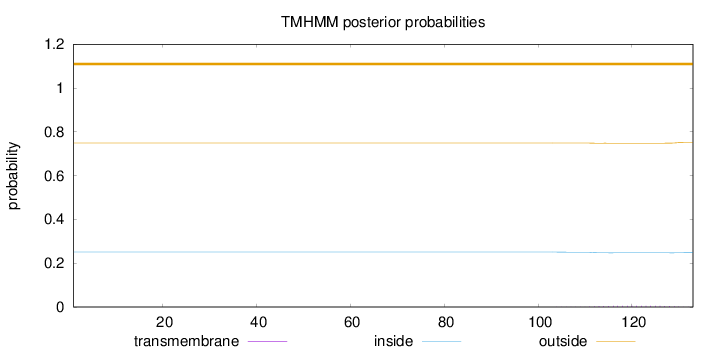
Length:
133
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09151
Exp number, first 60 AAs:
0
Total prob of N-in:
0.25112
outside
1 - 133
Population Genetic Test Statistics
Pi
278.390507
Theta
180.349747
Tajima's D
1.962997
CLR
0.522013
CSRT
0.87755612219389
Interpretation
Uncertain
