Gene
KWMTBOMO02659
Pre Gene Modal
BGIBMGA003446
Annotation
PREDICTED:_integrator_complex_subunit_6-A_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.533
Sequence
CDS
ATGACTATAATTGTATTTTTAATAGATACATCTGCTTCTATGAATCAAAGAGCTTATTTGGGCGGACGCCCAACATTACTCGATGTAGCCAAGGGCGCCGTGGAAACGTTCGTAAAGGTGCGTCAACGTTCCCCCGAAAGCAGAGGCGACAGGTATATGCTTTTAACTTTCGAAGAGCCACCCGCGAACATCAAGGCTGGTTGGAAAGAGAATCTGGCTACATTTATAAATGAATTAAAAAATCTACAATGTTTGGGTCTTACAACTATGGGTGCTGCCTTGAAACATGCATTTGATGTTTTGAATATTAACCGTATGCAGACTGGCATTGACACATATGGCCAAGGCAGATGCCCTTTCTACCTTGAGCCATCAGTTATAGTAGTAATCACAGATGGTGGCAAATTGTCAAGCAATGCTGGCATTCAGGAAGAGTTCAACTTGCCCATGAATTGCCCTATTCCCGGATCGGAGTTAACCCGGGAACCGTTTCGGTGGGATCAGCGTCTGTTCTCCCTTGTTTTGAGACTGGCCGGCACTCCGGCCGTAGAACGGGACACCGGCCTCGTGCCCTCCGATACGTCACCAATAGACGCCATGTGCGAAGTTACCGGAGGTCGTTCATACTGCATCACGTCGCACCGTATGTTGATGCAGTGCATCGACAGCTTAGTGCAGAAAGTGCAGAGCGGCGTCGTTATCAACTTTGAGAAGATCGGACCGGATCCGCCTCCGATCGACGGCGCCGCCGGCCGAGACGACCAGGACGAGGTGAATCACGACACCGAAAAGGAAGTGGACGCCGACGCCGACAGAAACGGGCGTTGGAACGGTGGCTCGTATCAGACCAGCGCGAATCAGAACCAGAGTGGCGGCGCACCGCAGGCTTGGCACTCGTGTCGTCGACTCATCTACGTGCCGCGGACCGTCGCCCAGAAAGGTTTCGCTGTAGGCTTCTGGCCAATACCGGAGTCGTTCAGATCGGATCCTAATGCACCCACTCTGCCACCTCGTTCAGCTCATCCAGTTGTGAAGTTCACATGTTCAAGCCAGGAGCCCATGGTCATCGACAATCTTCCCTTTGATAAGTACGAACTCGAGCCCAGTCCGTTGACACAATTCATACTCGCCAGAAAGCAACCAACAATTTGTTGGCAGGTGTTCGTGGCAAATAGCGGGTGTAAAACCTCAGAAATGAATCAACCGTTTGGTTACTTAAAAGCGTCAACGAACTTAACTTGTGTGAATCTGTTCGTACTACCCTACAACTACCCGATGCTATTGCCGTTGCTCGACGATTTGTTCAAAGTACATCGCTGCAAACCCACGCCGGAGTGGAAAATGGCCTTCCAGCACTATTTGGATCGCATGCCGGCCTACTATGTTGCGCCTCTACGAAGAGCCTTAACGAAAATGGGAGCATCACCACAGCTGCTTCAGAGCATCATACCCGACACTCAGGATAATTCTCTGAGTTTTTCCGTTCTGAACTATTTGAAAAGACTGAAGAATCAAGCGAAGATCGAATTTGAAAAGCTCTGCGCTGATGTTATTAATAAAGCCAATAGTAACAATCAAAAAGGAGCTGAAAGTGTTCGAGTAATGCCACGGTCGCCTCTCAAGAAAGACCTCACGACCCACCCGTGCCTTCAAGATAAATTTTCCGCCTTACGTGATCAACTCAATGAGTTTGGCGGCTTCGTGGTAGGACTCTCGCGGGGTCGCCATAAAAGCGGAGCCCACACATACCGTAATCCATTCGACATACAACGGAGAGATCTGTTGGATCAAGTGGTCCGAATGCGAGCCAATTTCCTGCAGCCCTCTCTCGCTCACACTCGTCTGTTAGACGAAGACAGCGCGCACTCCATGCCGGTCGCGCAGATGGGCAACTACCAGGAGTATCTGAAGCGGATGACGCCGCAGCTACGAGAGATCGAATCTCAGCCAGTTCGACAGCACATGTTCGGGAATCCATTCAAGATTGACAAACGCATGATGGTAGACGAAGCGGACATCGACCCCGTGGGGGGCGCGGTGCGAGGCGGGGCGGGGGGCGGGGCGGGCGGCAAGCGCGGCTCCGACCTCCCGCGGGTCACCCCACCTAAACGCAAGGCGGGACCTCTCCCGCAGCACGTGGTGGCGCGTCGTCCCTCCTCGTCCCCAGCCCCGCCGCTGTCCCCGCCCGCGGTCCCCCTGTCCGTCTCCCCGCAGCCGGCGCCGCTCGCGATTAGACGTCCGACATCACCACTAACGCCTTTGCCATCCGACTTGCCATCGGGACGACCACCCGAGGAGCCTCCAAACTTGGACTCGGGCGGGTCGGTGTGCGCGGCGCCGTGTCCGCCCGCGCCCCCCCTCGCGCCGCCCGCCAACCTGCCGCCCGTCGTCAAACCATTCCTCAACGGCGACGCGTACAGTAGTCCGTGCAAGGCTCCGCGGTCGCCGTCCCCGCCCAACGAGACGCGTTTGGACTCGCAGGAACTTCAGATCAACGCGATCCTGCAGGGGCTCGCTGCTCAGAACGACGCTCAGAACTCACGAGCGAAGAAATGGAAACGAAAAAGCGAGAGATTGGCAGCCCAGCCGCCCGCAGTGCCTCAACCTTCGCGGAAGGAGTTAGCCGACATACGTCGACACAACCTCGACGTGAGGGCGCAGGTGTACCGCGCCGTTAGACGACCCGGAAGAGATTTCACTAAACTGTTTGAGCACTTAAAACAATTGCAAGGTACTGTGGACATTAAAAAGGAAGTGTTACGTGATGTGATTAACGAATCCTTAAGGTTTAAAAGGAAGGCTCTAGCGAAATTACTAGAAGACTTCTTGGCCGGATTAGAGAAGAACGGAAGCAGCGCTTCCGGTACGATCAAGCGTTTGAACAACAGTGCGGTTTACAACAGTGGTGCGAATCATAGCTAG
Protein
MTIIVFLIDTSASMNQRAYLGGRPTLLDVAKGAVETFVKVRQRSPESRGDRYMLLTFEEPPANIKAGWKENLATFINELKNLQCLGLTTMGAALKHAFDVLNINRMQTGIDTYGQGRCPFYLEPSVIVVITDGGKLSSNAGIQEEFNLPMNCPIPGSELTREPFRWDQRLFSLVLRLAGTPAVERDTGLVPSDTSPIDAMCEVTGGRSYCITSHRMLMQCIDSLVQKVQSGVVINFEKIGPDPPPIDGAAGRDDQDEVNHDTEKEVDADADRNGRWNGGSYQTSANQNQSGGAPQAWHSCRRLIYVPRTVAQKGFAVGFWPIPESFRSDPNAPTLPPRSAHPVVKFTCSSQEPMVIDNLPFDKYELEPSPLTQFILARKQPTICWQVFVANSGCKTSEMNQPFGYLKASTNLTCVNLFVLPYNYPMLLPLLDDLFKVHRCKPTPEWKMAFQHYLDRMPAYYVAPLRRALTKMGASPQLLQSIIPDTQDNSLSFSVLNYLKRLKNQAKIEFEKLCADVINKANSNNQKGAESVRVMPRSPLKKDLTTHPCLQDKFSALRDQLNEFGGFVVGLSRGRHKSGAHTYRNPFDIQRRDLLDQVVRMRANFLQPSLAHTRLLDEDSAHSMPVAQMGNYQEYLKRMTPQLREIESQPVRQHMFGNPFKIDKRMMVDEADIDPVGGAVRGGAGGGAGGKRGSDLPRVTPPKRKAGPLPQHVVARRPSSSPAPPLSPPAVPLSVSPQPAPLAIRRPTSPLTPLPSDLPSGRPPEEPPNLDSGGSVCAAPCPPAPPLAPPANLPPVVKPFLNGDAYSSPCKAPRSPSPPNETRLDSQELQINAILQGLAAQNDAQNSRAKKWKRKSERLAAQPPAVPQPSRKELADIRRHNLDVRAQVYRAVRRPGRDFTKLFEHLKQLQGTVDIKKEVLRDVINESLRFKRKALAKLLEDFLAGLEKNGSSASGTIKRLNNSAVYNSGANHS
Summary
Uniprot
H9J1Q9
A0A212F3P6
A0A194RH68
A0A194PP81
A0A0L7LLX1
A0A2A3ECW9
+ More
A0A2J7QX31 A0A067RLG8 A0A088ARH9 A0A0L7RIR1 A0A3L8DRQ1 A0A154P650 A0A195FF83 A0A158NGN0 F4WZV8 E9ILA0 E2BAQ9 E2ACC5 A0A0C9RLZ6 A0A2H1W647 A0A151XAG9 A0A151ICF9 A0A224X688 A0A023F2P8 A0A1B6HDR4 A0A195B7U0 K7IUG7 A0A0M9A6H7 A0A232F8S6 A0A2P8Y3S5 A0A0P5YWA1 A0A0P5G8L5 A0A0P5XVT7 A0A0P6II59 A0A0P6FDT2 A0A0P5S4G6 A0A0P5MRN6 A0A0P6EE64 A0A0P5WBK5 A0A0P5AYY5 A0A0P5NIA8 A0A0P5HQJ3 A0A0P5VEA5 A0A0P5WQY5 A0A0P5AZS3 A0A0P4Z946 A0A0P5GWC9 A0A0P6DEA1 A0A0P5ES36 A0A0P5Q5V3 A0A0P4YYW5 A0A0P5HI02 A0A0P5S5B0 A0A0N7ZZH1 A0A0P5Q153 A0A0P5ABF1 A0A0P5JL65 A0A0P6FAJ8 A0A0P5ERA7 A0A0P5AIL2 A0A0P5BFG6 A0A0P5F9N1 A0A0P5X082 A0A0P5B4X1 A0A0P6FS72 A0A0P5W0K7 A0A0P5R2W7 A0A0P5V4P0 A0A0P5V1B0 A0A0P5V701 A0A0P5UZU4 A0A0P5V412 A0A0P4ZP96 A0A0N8CNB4 A0A0P4ZFE1 A0A0P5Y7I1 D2A291 A0A2H8TKG4 J9JY41 A0A2S2NRC2 A0A0P5Y2C0 A0A0P5Y2B6 T1J717 E9H4B7 U4U667 A0A0P5B6M3
A0A2J7QX31 A0A067RLG8 A0A088ARH9 A0A0L7RIR1 A0A3L8DRQ1 A0A154P650 A0A195FF83 A0A158NGN0 F4WZV8 E9ILA0 E2BAQ9 E2ACC5 A0A0C9RLZ6 A0A2H1W647 A0A151XAG9 A0A151ICF9 A0A224X688 A0A023F2P8 A0A1B6HDR4 A0A195B7U0 K7IUG7 A0A0M9A6H7 A0A232F8S6 A0A2P8Y3S5 A0A0P5YWA1 A0A0P5G8L5 A0A0P5XVT7 A0A0P6II59 A0A0P6FDT2 A0A0P5S4G6 A0A0P5MRN6 A0A0P6EE64 A0A0P5WBK5 A0A0P5AYY5 A0A0P5NIA8 A0A0P5HQJ3 A0A0P5VEA5 A0A0P5WQY5 A0A0P5AZS3 A0A0P4Z946 A0A0P5GWC9 A0A0P6DEA1 A0A0P5ES36 A0A0P5Q5V3 A0A0P4YYW5 A0A0P5HI02 A0A0P5S5B0 A0A0N7ZZH1 A0A0P5Q153 A0A0P5ABF1 A0A0P5JL65 A0A0P6FAJ8 A0A0P5ERA7 A0A0P5AIL2 A0A0P5BFG6 A0A0P5F9N1 A0A0P5X082 A0A0P5B4X1 A0A0P6FS72 A0A0P5W0K7 A0A0P5R2W7 A0A0P5V4P0 A0A0P5V1B0 A0A0P5V701 A0A0P5UZU4 A0A0P5V412 A0A0P4ZP96 A0A0N8CNB4 A0A0P4ZFE1 A0A0P5Y7I1 D2A291 A0A2H8TKG4 J9JY41 A0A2S2NRC2 A0A0P5Y2C0 A0A0P5Y2B6 T1J717 E9H4B7 U4U667 A0A0P5B6M3
Pubmed
EMBL
BABH01008184
BABH01008185
BABH01008186
AGBW02010519
OWR48355.1
KQ460207
+ More
KPJ16670.1 KQ459597 KPI94788.1 JTDY01000647 KOB76349.1 KZ288282 PBC29577.1 NEVH01009398 PNF33141.1 KK852554 KDR21465.1 KQ414584 KOC70626.1 QOIP01000005 RLU22529.1 KQ434826 KZC07415.1 KQ981625 KYN39048.1 ADTU01015147 ADTU01015148 GL888480 EGI60248.1 GL764074 EFZ18638.1 GL446831 EFN87194.1 GL438491 EFN68915.1 GBYB01014392 JAG84159.1 ODYU01006551 SOQ48493.1 KQ982339 KYQ57356.1 KQ978053 KYM97765.1 GFTR01008439 JAW07987.1 GBBI01003112 JAC15600.1 GECU01034812 JAS72894.1 KQ976574 KYM80312.1 AAZX01002634 KQ435726 KOX78227.1 NNAY01000641 OXU27244.1 PYGN01000957 PSN38922.1 GDIP01052394 JAM51321.1 GDIQ01252135 JAJ99589.1 GDIP01066581 JAM37134.1 GDIP01083633 GDIQ01010655 JAM20082.1 JAN84082.1 GDIQ01049161 JAN45576.1 GDIQ01096926 JAL54800.1 GDIQ01152266 JAK99459.1 GDIQ01078230 JAN16507.1 GDIP01101531 JAM02184.1 GDIP01192072 JAJ31330.1 GDIQ01152264 JAK99461.1 GDIQ01224641 JAK27084.1 GDIP01101529 JAM02186.1 GDIP01083632 JAM20083.1 GDIP01216217 GDIP01192070 GDIQ01208044 GDIQ01172648 GDIQ01106231 GDIP01101491 LRGB01001728 JAJ31332.1 JAK43681.1 KZS10731.1 GDIP01216218 GDIP01192071 GDIQ01066945 JAJ07184.1 JAN27792.1 GDIQ01235533 JAK16192.1 GDIQ01080187 JAN14550.1 GDIQ01265521 JAJ86203.1 GDIQ01128529 JAL23197.1 GDIP01220875 JAJ02527.1 GDIQ01233182 JAK18543.1 GDIQ01098583 JAL53143.1 GDIP01197369 JAJ26033.1 GDIQ01127180 JAL24546.1 GDIP01201471 JAJ21931.1 GDIQ01196791 JAK54934.1 GDIQ01050560 JAN44177.1 GDIQ01265869 JAJ85855.1 GDIP01201472 JAJ21930.1 GDIP01189614 JAJ33788.1 GDIQ01265868 JAJ85856.1 GDIP01079284 JAM24431.1 GDIP01189616 JAJ33786.1 GDIP01122293 GDIQ01043846 JAL81421.1 JAN50891.1 GDIP01107035 JAL96679.1 GDIQ01106232 JAL45494.1 GDIP01104443 JAL99271.1 GDIP01107033 JAL96681.1 GDIP01104441 JAL99273.1 GDIP01107034 JAL96680.1 GDIP01104442 JAL99272.1 GDIP01223109 JAJ00293.1 GDIP01114935 JAL88779.1 GDIP01214109 JAJ09293.1 GDIP01075035 JAM28680.1 KQ971338 EFA02055.1 GFXV01002820 MBW14625.1 ABLF02040117 GGMR01007085 MBY19704.1 GDIP01063938 JAM39777.1 GDIP01063939 JAM39776.1 JH431901 GL732591 EFX73352.1 KB631697 ERL85445.1 GDIP01189615 JAJ33787.1
KPJ16670.1 KQ459597 KPI94788.1 JTDY01000647 KOB76349.1 KZ288282 PBC29577.1 NEVH01009398 PNF33141.1 KK852554 KDR21465.1 KQ414584 KOC70626.1 QOIP01000005 RLU22529.1 KQ434826 KZC07415.1 KQ981625 KYN39048.1 ADTU01015147 ADTU01015148 GL888480 EGI60248.1 GL764074 EFZ18638.1 GL446831 EFN87194.1 GL438491 EFN68915.1 GBYB01014392 JAG84159.1 ODYU01006551 SOQ48493.1 KQ982339 KYQ57356.1 KQ978053 KYM97765.1 GFTR01008439 JAW07987.1 GBBI01003112 JAC15600.1 GECU01034812 JAS72894.1 KQ976574 KYM80312.1 AAZX01002634 KQ435726 KOX78227.1 NNAY01000641 OXU27244.1 PYGN01000957 PSN38922.1 GDIP01052394 JAM51321.1 GDIQ01252135 JAJ99589.1 GDIP01066581 JAM37134.1 GDIP01083633 GDIQ01010655 JAM20082.1 JAN84082.1 GDIQ01049161 JAN45576.1 GDIQ01096926 JAL54800.1 GDIQ01152266 JAK99459.1 GDIQ01078230 JAN16507.1 GDIP01101531 JAM02184.1 GDIP01192072 JAJ31330.1 GDIQ01152264 JAK99461.1 GDIQ01224641 JAK27084.1 GDIP01101529 JAM02186.1 GDIP01083632 JAM20083.1 GDIP01216217 GDIP01192070 GDIQ01208044 GDIQ01172648 GDIQ01106231 GDIP01101491 LRGB01001728 JAJ31332.1 JAK43681.1 KZS10731.1 GDIP01216218 GDIP01192071 GDIQ01066945 JAJ07184.1 JAN27792.1 GDIQ01235533 JAK16192.1 GDIQ01080187 JAN14550.1 GDIQ01265521 JAJ86203.1 GDIQ01128529 JAL23197.1 GDIP01220875 JAJ02527.1 GDIQ01233182 JAK18543.1 GDIQ01098583 JAL53143.1 GDIP01197369 JAJ26033.1 GDIQ01127180 JAL24546.1 GDIP01201471 JAJ21931.1 GDIQ01196791 JAK54934.1 GDIQ01050560 JAN44177.1 GDIQ01265869 JAJ85855.1 GDIP01201472 JAJ21930.1 GDIP01189614 JAJ33788.1 GDIQ01265868 JAJ85856.1 GDIP01079284 JAM24431.1 GDIP01189616 JAJ33786.1 GDIP01122293 GDIQ01043846 JAL81421.1 JAN50891.1 GDIP01107035 JAL96679.1 GDIQ01106232 JAL45494.1 GDIP01104443 JAL99271.1 GDIP01107033 JAL96681.1 GDIP01104441 JAL99273.1 GDIP01107034 JAL96680.1 GDIP01104442 JAL99272.1 GDIP01223109 JAJ00293.1 GDIP01114935 JAL88779.1 GDIP01214109 JAJ09293.1 GDIP01075035 JAM28680.1 KQ971338 EFA02055.1 GFXV01002820 MBW14625.1 ABLF02040117 GGMR01007085 MBY19704.1 GDIP01063938 JAM39777.1 GDIP01063939 JAM39776.1 JH431901 GL732591 EFX73352.1 KB631697 ERL85445.1 GDIP01189615 JAJ33787.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000242457
+ More
UP000235965 UP000027135 UP000005203 UP000053825 UP000279307 UP000076502 UP000078541 UP000005205 UP000007755 UP000008237 UP000000311 UP000075809 UP000078542 UP000078540 UP000002358 UP000053105 UP000215335 UP000245037 UP000076858 UP000007266 UP000007819 UP000000305 UP000030742
UP000235965 UP000027135 UP000005203 UP000053825 UP000279307 UP000076502 UP000078541 UP000005205 UP000007755 UP000008237 UP000000311 UP000075809 UP000078542 UP000078540 UP000002358 UP000053105 UP000215335 UP000245037 UP000076858 UP000007266 UP000007819 UP000000305 UP000030742
PRIDE
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
H9J1Q9
A0A212F3P6
A0A194RH68
A0A194PP81
A0A0L7LLX1
A0A2A3ECW9
+ More
A0A2J7QX31 A0A067RLG8 A0A088ARH9 A0A0L7RIR1 A0A3L8DRQ1 A0A154P650 A0A195FF83 A0A158NGN0 F4WZV8 E9ILA0 E2BAQ9 E2ACC5 A0A0C9RLZ6 A0A2H1W647 A0A151XAG9 A0A151ICF9 A0A224X688 A0A023F2P8 A0A1B6HDR4 A0A195B7U0 K7IUG7 A0A0M9A6H7 A0A232F8S6 A0A2P8Y3S5 A0A0P5YWA1 A0A0P5G8L5 A0A0P5XVT7 A0A0P6II59 A0A0P6FDT2 A0A0P5S4G6 A0A0P5MRN6 A0A0P6EE64 A0A0P5WBK5 A0A0P5AYY5 A0A0P5NIA8 A0A0P5HQJ3 A0A0P5VEA5 A0A0P5WQY5 A0A0P5AZS3 A0A0P4Z946 A0A0P5GWC9 A0A0P6DEA1 A0A0P5ES36 A0A0P5Q5V3 A0A0P4YYW5 A0A0P5HI02 A0A0P5S5B0 A0A0N7ZZH1 A0A0P5Q153 A0A0P5ABF1 A0A0P5JL65 A0A0P6FAJ8 A0A0P5ERA7 A0A0P5AIL2 A0A0P5BFG6 A0A0P5F9N1 A0A0P5X082 A0A0P5B4X1 A0A0P6FS72 A0A0P5W0K7 A0A0P5R2W7 A0A0P5V4P0 A0A0P5V1B0 A0A0P5V701 A0A0P5UZU4 A0A0P5V412 A0A0P4ZP96 A0A0N8CNB4 A0A0P4ZFE1 A0A0P5Y7I1 D2A291 A0A2H8TKG4 J9JY41 A0A2S2NRC2 A0A0P5Y2C0 A0A0P5Y2B6 T1J717 E9H4B7 U4U667 A0A0P5B6M3
A0A2J7QX31 A0A067RLG8 A0A088ARH9 A0A0L7RIR1 A0A3L8DRQ1 A0A154P650 A0A195FF83 A0A158NGN0 F4WZV8 E9ILA0 E2BAQ9 E2ACC5 A0A0C9RLZ6 A0A2H1W647 A0A151XAG9 A0A151ICF9 A0A224X688 A0A023F2P8 A0A1B6HDR4 A0A195B7U0 K7IUG7 A0A0M9A6H7 A0A232F8S6 A0A2P8Y3S5 A0A0P5YWA1 A0A0P5G8L5 A0A0P5XVT7 A0A0P6II59 A0A0P6FDT2 A0A0P5S4G6 A0A0P5MRN6 A0A0P6EE64 A0A0P5WBK5 A0A0P5AYY5 A0A0P5NIA8 A0A0P5HQJ3 A0A0P5VEA5 A0A0P5WQY5 A0A0P5AZS3 A0A0P4Z946 A0A0P5GWC9 A0A0P6DEA1 A0A0P5ES36 A0A0P5Q5V3 A0A0P4YYW5 A0A0P5HI02 A0A0P5S5B0 A0A0N7ZZH1 A0A0P5Q153 A0A0P5ABF1 A0A0P5JL65 A0A0P6FAJ8 A0A0P5ERA7 A0A0P5AIL2 A0A0P5BFG6 A0A0P5F9N1 A0A0P5X082 A0A0P5B4X1 A0A0P6FS72 A0A0P5W0K7 A0A0P5R2W7 A0A0P5V4P0 A0A0P5V1B0 A0A0P5V701 A0A0P5UZU4 A0A0P5V412 A0A0P4ZP96 A0A0N8CNB4 A0A0P4ZFE1 A0A0P5Y7I1 D2A291 A0A2H8TKG4 J9JY41 A0A2S2NRC2 A0A0P5Y2C0 A0A0P5Y2B6 T1J717 E9H4B7 U4U667 A0A0P5B6M3
Ontologies
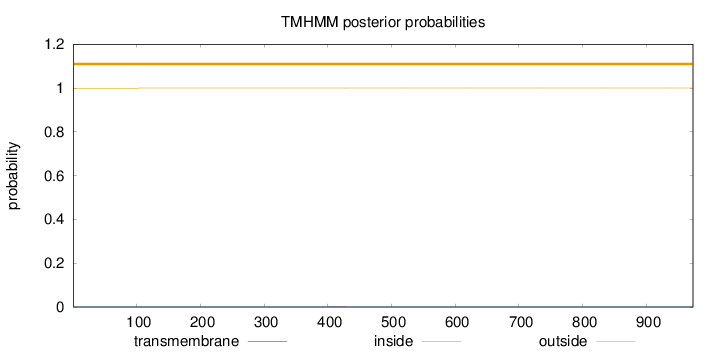
Topology
Length:
973
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01142
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.00053
outside
1 - 973
Population Genetic Test Statistics
Pi
260.40174
Theta
189.133289
Tajima's D
1.162423
CLR
0.048014
CSRT
0.696115194240288
Interpretation
Uncertain
