Pre Gene Modal
BGIBMGA002654
Annotation
putative_ATP-dependent_RNA_helicase_DDX51_[Operophtera_brumata]
Full name
Probable ATP-dependent RNA helicase Dbp73D
Location in the cell
Nuclear Reliability : 2.475
Sequence
CDS
ATGGATTTATTTATTATTAACAGATATAATGACACAAATGATGATACACATATAATAGAACAAGAAGCAAAACGCCTAGAACAATTGAAGCAAAGAATTGAAGAACGTAGAAAAGCATTTTCAGTAAAAAAATCGTCTGATGTCATTACTTTGAAAGAAGAAAAACAAACTGAAATTTTACCTGACCTTCCCGTTTCTACTAATGATGATTTGCCAACGGAAATACCAAAACCAAATACACCTCCAAAACCAAAAGAAAAAATTTATGCTGAAAAACAAAAAGAATTTAAAGTTCTTGGATCAAGTGACTTTCAAAAAAAATCTAAGGTGCAGAGAATATTACCATACTGGCTATCGCATCCTACAGTTGTATCAAGAGATTTAAATGAACTATCTTGTCCAGTTGAAGAACAAGAATGGCTACACCCAGTAATAAAGTCTACTTTATTGAGCGAAGGCATAACAAAACTATTTCCAGTTCAAAATCAAGTGGTACCATTCATTTTAAATGAGCATACATTGCCAAAATTATTTCAACCTCGTGACGTTTGCATCTCAGCACCAACAGGAAGTGGCAAAACACTGTCATTTGTCATACCTATAGTGCAGCTTCTAATGAAAGATGTGGGTTGCAAAATACGAGCACTTGTGGTATTACCAGTACAAGAGTTAGCTGCTCAAGTTGCTAAAGTATTTAAGAAGTATTGCAATAGAACTCATCTTAAAGTTGCTCTCTTGAGTGGTTCAGTTCCCTTTCAACAGGAACAGCAAGAGCTTGTCAGATTTACTGAATCAGTAGGATGGGTCAGTAAAGTAGAAATCATTGTATGTACAGCAGGAAGATTGGTTGAGCACTTAAAGACTACAGAAGGTTTCAATTTACAACATTTAAAGTTTTTAGTTATTGATGAAGCTGACCGAATTATGGATAACATACAAAATGATTGGCTCTACCATGTTGATCATCATATCAAACAAGGGAGTGAATTGATGACAGGGAAAGTTCAGAGTTTAAATTGGAAAAATGTGAGTAAACAAAAATTAGCACCCCAAAAATTACTCTTCTCTGCCACATTATCCCAAGATCCAGAGAAGTTGGAACAGTGGGGGCTATTTCAACCAAAATTGTTTTCTGCTGCCACTAATGAATGTTTTGAAGATGAAGACCAAATCAGAAAATATGCTACACCTGCAGAACTTAAGGAACACTATGTTGTATGTGAAGCAGAGAACAAACCTCTTGTACTTTATAATTTTTTAGTAGAACAAAAATGGGATAAAGTATTATGTTTTACAAATGCTGCTGAAACAACTCACAGGCTAACTGTACTTTTAAACACATGGGCTTGTAAAGGATTAAAAGTAGCTGAATTGTCAGCCGCTTTAGATAGATCAATGAGGGAAAATGTTTTAAAGAAATTCATGCTCTCTGAAATTGATGTGTTAATCAGCACTGATGCATTAGCCAGAGGTATCGATATACCAGATTGCAACTACGTAATATCTTATGATCCTCCACGGAACATAAAAACATACATACATCGAGTTGGTAGAACAGGAAGAGCTGGAAAGATTGGTAAAGCAGTAACTGTGCTATTGCCAAGTCAAATCAGCTTATTTAAGGATCTTATAAAAGCAGGAGGAAAAGAGGAAATTCCGGAAATTAAAATTAATACTGATGTAATTGATCGTTTATTCGAAAGTTATGCATCTGCCATGAACACAACGAGAGAAAAAATAAATTCAGAGCTAAACTTGAAAATTAAAAACTCAATAGAAATTAAAAGAAGTGCTAAAAGTAAAATTCAAAAGAGAAAAAAATAG
Protein
MDLFIINRYNDTNDDTHIIEQEAKRLEQLKQRIEERRKAFSVKKSSDVITLKEEKQTEILPDLPVSTNDDLPTEIPKPNTPPKPKEKIYAEKQKEFKVLGSSDFQKKSKVQRILPYWLSHPTVVSRDLNELSCPVEEQEWLHPVIKSTLLSEGITKLFPVQNQVVPFILNEHTLPKLFQPRDVCISAPTGSGKTLSFVIPIVQLLMKDVGCKIRALVVLPVQELAAQVAKVFKKYCNRTHLKVALLSGSVPFQQEQQELVRFTESVGWVSKVEIIVCTAGRLVEHLKTTEGFNLQHLKFLVIDEADRIMDNIQNDWLYHVDHHIKQGSELMTGKVQSLNWKNVSKQKLAPQKLLFSATLSQDPEKLEQWGLFQPKLFSAATNECFEDEDQIRKYATPAELKEHYVVCEAENKPLVLYNFLVEQKWDKVLCFTNAAETTHRLTVLLNTWACKGLKVAELSAALDRSMRENVLKKFMLSEIDVLISTDALARGIDIPDCNYVISYDPPRNIKTYIHRVGRTGRAGKIGKAVTVLLPSQISLFKDLIKAGGKEEIPEIKINTDVIDRLFESYASAMNTTREKINSELNLKIKNSIEIKRSAKSKIQKRKK
Summary
Description
ATP-binding RNA helicase involved in the biogenesis of 60S ribosomal subunits.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family. DDX51/DBP6 subfamily.
Belongs to the DEAD box helicase family. DDX51/DBP6 subfamily.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Ribosome biogenesis
RNA-binding
rRNA processing
Feature
chain Probable ATP-dependent RNA helicase Dbp73D
Uniprot
H9IZG9
A0A2H1WBW1
A0A2A4K6G6
A0A2W1BH27
A0A0L7LVF8
A0A194RGE4
+ More
A0A194PMW9 A0A212FJV1 D6WD90 A0A1Y1M9I5 A0A336KF26 A0A336MJC5 A0A2J7PJ57 W8BUG2 A0A0A1WSX0 A0A084W143 A0A0L0CB09 A0A067QQD0 A0A1I8NT42 A0A1I8NT35 A0A034V452 N6TWN3 A0A1Q3F1K4 A0A0K8UPS4 E0VWC9 A0A1Q3F1X7 A0A182N9A6 A0A1I8M450 A0A2M4APT5 A0A1I8MVI4 A0A1A9XQZ6 W5JV16 A0A2M4ANH7 A0A1A9UP15 A0A182JFD2 A0A1B0DIE3 A0A1A9Z922 A0A182PIJ1 A0A182TEN0 A0A182WWI2 B4LG52 A0A182I6R2 Q7QIL5 A0A182L3N3 A0A0P6IVI1 A0A1B0G7J1 A0A182WBI4 A0A182MFL8 B0W5M0 A0A1B6GYE8 B4KXL0 A0A1B0C2P6 A0A0A9Z2H7 B4MM57 A0A182FEM3 A0A069DV92 A0A182JRN8 A0A182Y2B7 A0A1S4GTP2 A0A146L1M0 A0A0V0G5F8 B4IXF5 P26802 A0A1W4UGI4 A0A1A9W4X8 A0A3B0KLV4 A0A3B0KUD8 B4PKA4 B3NDI1 A0A182VAG1 A0A182RJL1 A0A224X9K7 A0A182Q8N8 B3M4E5 B4QMY0 Q29DT5 A0A023EZG8 A0A1B6C6W5 B4HK61 T1PJQ9 A0A0M5J467 A0A2K6P3N6 A0A2R2MRE2 A0A2P8ZHG2 G1M4Z3 A0A1U7R9Y5 A0A2K5UU65 A0A1J1IBZ4 A0A2K5IPK8 A0A287BGR7 A0A151N4A6 A0A091N2K9 D2HLG6 A0A2K6BYN8 A0A2K5RE81 A0A2K5M739 G7PJH3 A0A2K5RE66 A0A2K6FP00
A0A194PMW9 A0A212FJV1 D6WD90 A0A1Y1M9I5 A0A336KF26 A0A336MJC5 A0A2J7PJ57 W8BUG2 A0A0A1WSX0 A0A084W143 A0A0L0CB09 A0A067QQD0 A0A1I8NT42 A0A1I8NT35 A0A034V452 N6TWN3 A0A1Q3F1K4 A0A0K8UPS4 E0VWC9 A0A1Q3F1X7 A0A182N9A6 A0A1I8M450 A0A2M4APT5 A0A1I8MVI4 A0A1A9XQZ6 W5JV16 A0A2M4ANH7 A0A1A9UP15 A0A182JFD2 A0A1B0DIE3 A0A1A9Z922 A0A182PIJ1 A0A182TEN0 A0A182WWI2 B4LG52 A0A182I6R2 Q7QIL5 A0A182L3N3 A0A0P6IVI1 A0A1B0G7J1 A0A182WBI4 A0A182MFL8 B0W5M0 A0A1B6GYE8 B4KXL0 A0A1B0C2P6 A0A0A9Z2H7 B4MM57 A0A182FEM3 A0A069DV92 A0A182JRN8 A0A182Y2B7 A0A1S4GTP2 A0A146L1M0 A0A0V0G5F8 B4IXF5 P26802 A0A1W4UGI4 A0A1A9W4X8 A0A3B0KLV4 A0A3B0KUD8 B4PKA4 B3NDI1 A0A182VAG1 A0A182RJL1 A0A224X9K7 A0A182Q8N8 B3M4E5 B4QMY0 Q29DT5 A0A023EZG8 A0A1B6C6W5 B4HK61 T1PJQ9 A0A0M5J467 A0A2K6P3N6 A0A2R2MRE2 A0A2P8ZHG2 G1M4Z3 A0A1U7R9Y5 A0A2K5UU65 A0A1J1IBZ4 A0A2K5IPK8 A0A287BGR7 A0A151N4A6 A0A091N2K9 D2HLG6 A0A2K6BYN8 A0A2K5RE81 A0A2K5M739 G7PJH3 A0A2K5RE66 A0A2K6FP00
EC Number
3.6.4.13
Pubmed
19121390
28756777
26227816
26354079
22118469
18362917
+ More
19820115 28004739 24495485 25830018 24438588 26108605 24845553 25348373 23537049 20566863 25315136 20920257 23761445 17994087 12364791 20966253 26999592 25401762 26334808 25244985 26823975 1620603 10731132 12537572 12537569 17550304 22936249 15632085 25474469 25362486 29403074 20010809 22293439 22002653
19820115 28004739 24495485 25830018 24438588 26108605 24845553 25348373 23537049 20566863 25315136 20920257 23761445 17994087 12364791 20966253 26999592 25401762 26334808 25244985 26823975 1620603 10731132 12537572 12537569 17550304 22936249 15632085 25474469 25362486 29403074 20010809 22293439 22002653
EMBL
BABH01014116
BABH01014117
ODYU01007628
SOQ50575.1
NWSH01000078
PCG79831.1
+ More
KZ150098 PZC73571.1 JTDY01000024 KOB79364.1 KQ460207 KPJ16657.1 KQ459597 KPI94776.1 AGBW02008206 OWR54011.1 KQ971321 EEZ99527.1 GEZM01036946 JAV82323.1 UFQS01000244 UFQT01000244 SSX01935.1 SSX22312.1 UFQT01001136 SSX29079.1 NEVH01024950 PNF16375.1 GAMC01013786 JAB92769.1 GBXI01012774 JAD01518.1 ATLV01019208 KE525264 KFB43937.1 JRES01000665 KNC29431.1 KK853579 KDR06549.1 GAKP01021678 JAC37274.1 APGK01058443 KB741288 KB632370 ENN70712.1 ERL93709.1 GFDL01013605 JAV21440.1 GDHF01023801 JAI28513.1 DS235818 EEB17685.1 GFDL01013482 JAV21563.1 GGFK01009459 MBW42780.1 ADMH02000413 ETN66624.1 GGFK01009010 MBW42331.1 AJVK01034367 CH940647 EDW69360.1 APCN01003693 AAAB01008807 EAA04138.4 GDUN01000083 JAN95836.1 CCAG010022573 AXCM01000934 DS231843 EDS35503.1 GECZ01002324 JAS67445.1 CH933809 EDW18696.1 JXJN01024604 GBHO01004970 GBHO01004969 GBRD01008059 JAG38634.1 JAG38635.1 JAG57762.1 CH963847 EDW73066.1 GBGD01000901 JAC87988.1 GDHC01016780 JAQ01849.1 GECL01002846 JAP03278.1 CH916366 EDV96392.1 M74824 AE014296 AF132173 BT010045 OUUW01000012 SPP87559.1 SPP87558.1 CM000159 EDW94802.1 CH954178 EDV52043.1 GFTR01007389 JAW09037.1 AXCN02000558 CH902618 EDV40439.1 CM000363 CM002912 EDX10772.1 KMZ00101.1 CH379070 EAL30329.2 GBBI01004079 JAC14633.1 GEDC01028055 JAS09243.1 CH480815 EDW41802.1 KA649022 AFP63651.1 CP012525 ALC43426.1 ALC44899.1 ALC44906.1 PYGN01000057 PSN55931.1 ACTA01027180 ACTA01035180 AQIA01013500 CVRI01000047 CRK97797.1 AEMK02000096 AKHW03004053 KYO31622.1 KL375005 KFP83134.1 GL193005 EFB16388.1 CM001286 EHH66849.1
KZ150098 PZC73571.1 JTDY01000024 KOB79364.1 KQ460207 KPJ16657.1 KQ459597 KPI94776.1 AGBW02008206 OWR54011.1 KQ971321 EEZ99527.1 GEZM01036946 JAV82323.1 UFQS01000244 UFQT01000244 SSX01935.1 SSX22312.1 UFQT01001136 SSX29079.1 NEVH01024950 PNF16375.1 GAMC01013786 JAB92769.1 GBXI01012774 JAD01518.1 ATLV01019208 KE525264 KFB43937.1 JRES01000665 KNC29431.1 KK853579 KDR06549.1 GAKP01021678 JAC37274.1 APGK01058443 KB741288 KB632370 ENN70712.1 ERL93709.1 GFDL01013605 JAV21440.1 GDHF01023801 JAI28513.1 DS235818 EEB17685.1 GFDL01013482 JAV21563.1 GGFK01009459 MBW42780.1 ADMH02000413 ETN66624.1 GGFK01009010 MBW42331.1 AJVK01034367 CH940647 EDW69360.1 APCN01003693 AAAB01008807 EAA04138.4 GDUN01000083 JAN95836.1 CCAG010022573 AXCM01000934 DS231843 EDS35503.1 GECZ01002324 JAS67445.1 CH933809 EDW18696.1 JXJN01024604 GBHO01004970 GBHO01004969 GBRD01008059 JAG38634.1 JAG38635.1 JAG57762.1 CH963847 EDW73066.1 GBGD01000901 JAC87988.1 GDHC01016780 JAQ01849.1 GECL01002846 JAP03278.1 CH916366 EDV96392.1 M74824 AE014296 AF132173 BT010045 OUUW01000012 SPP87559.1 SPP87558.1 CM000159 EDW94802.1 CH954178 EDV52043.1 GFTR01007389 JAW09037.1 AXCN02000558 CH902618 EDV40439.1 CM000363 CM002912 EDX10772.1 KMZ00101.1 CH379070 EAL30329.2 GBBI01004079 JAC14633.1 GEDC01028055 JAS09243.1 CH480815 EDW41802.1 KA649022 AFP63651.1 CP012525 ALC43426.1 ALC44899.1 ALC44906.1 PYGN01000057 PSN55931.1 ACTA01027180 ACTA01035180 AQIA01013500 CVRI01000047 CRK97797.1 AEMK02000096 AKHW03004053 KYO31622.1 KL375005 KFP83134.1 GL193005 EFB16388.1 CM001286 EHH66849.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000235965 UP000030765 UP000037069 UP000027135 UP000095300 UP000019118 UP000030742 UP000009046 UP000075884 UP000095301 UP000092443 UP000000673 UP000078200 UP000075880 UP000092462 UP000092445 UP000075885 UP000075902 UP000076407 UP000008792 UP000075840 UP000007062 UP000075882 UP000092444 UP000075920 UP000075883 UP000002320 UP000009192 UP000092460 UP000007798 UP000069272 UP000075881 UP000076408 UP000001070 UP000000803 UP000192221 UP000091820 UP000268350 UP000002282 UP000008711 UP000075903 UP000075900 UP000075886 UP000007801 UP000000304 UP000001819 UP000001292 UP000092553 UP000233200 UP000085678 UP000245037 UP000008912 UP000189705 UP000233100 UP000183832 UP000233080 UP000008227 UP000050525 UP000233120 UP000233040 UP000233060 UP000009130 UP000233160
UP000007266 UP000235965 UP000030765 UP000037069 UP000027135 UP000095300 UP000019118 UP000030742 UP000009046 UP000075884 UP000095301 UP000092443 UP000000673 UP000078200 UP000075880 UP000092462 UP000092445 UP000075885 UP000075902 UP000076407 UP000008792 UP000075840 UP000007062 UP000075882 UP000092444 UP000075920 UP000075883 UP000002320 UP000009192 UP000092460 UP000007798 UP000069272 UP000075881 UP000076408 UP000001070 UP000000803 UP000192221 UP000091820 UP000268350 UP000002282 UP000008711 UP000075903 UP000075900 UP000075886 UP000007801 UP000000304 UP000001819 UP000001292 UP000092553 UP000233200 UP000085678 UP000245037 UP000008912 UP000189705 UP000233100 UP000183832 UP000233080 UP000008227 UP000050525 UP000233120 UP000233040 UP000233060 UP000009130 UP000233160
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9IZG9
A0A2H1WBW1
A0A2A4K6G6
A0A2W1BH27
A0A0L7LVF8
A0A194RGE4
+ More
A0A194PMW9 A0A212FJV1 D6WD90 A0A1Y1M9I5 A0A336KF26 A0A336MJC5 A0A2J7PJ57 W8BUG2 A0A0A1WSX0 A0A084W143 A0A0L0CB09 A0A067QQD0 A0A1I8NT42 A0A1I8NT35 A0A034V452 N6TWN3 A0A1Q3F1K4 A0A0K8UPS4 E0VWC9 A0A1Q3F1X7 A0A182N9A6 A0A1I8M450 A0A2M4APT5 A0A1I8MVI4 A0A1A9XQZ6 W5JV16 A0A2M4ANH7 A0A1A9UP15 A0A182JFD2 A0A1B0DIE3 A0A1A9Z922 A0A182PIJ1 A0A182TEN0 A0A182WWI2 B4LG52 A0A182I6R2 Q7QIL5 A0A182L3N3 A0A0P6IVI1 A0A1B0G7J1 A0A182WBI4 A0A182MFL8 B0W5M0 A0A1B6GYE8 B4KXL0 A0A1B0C2P6 A0A0A9Z2H7 B4MM57 A0A182FEM3 A0A069DV92 A0A182JRN8 A0A182Y2B7 A0A1S4GTP2 A0A146L1M0 A0A0V0G5F8 B4IXF5 P26802 A0A1W4UGI4 A0A1A9W4X8 A0A3B0KLV4 A0A3B0KUD8 B4PKA4 B3NDI1 A0A182VAG1 A0A182RJL1 A0A224X9K7 A0A182Q8N8 B3M4E5 B4QMY0 Q29DT5 A0A023EZG8 A0A1B6C6W5 B4HK61 T1PJQ9 A0A0M5J467 A0A2K6P3N6 A0A2R2MRE2 A0A2P8ZHG2 G1M4Z3 A0A1U7R9Y5 A0A2K5UU65 A0A1J1IBZ4 A0A2K5IPK8 A0A287BGR7 A0A151N4A6 A0A091N2K9 D2HLG6 A0A2K6BYN8 A0A2K5RE81 A0A2K5M739 G7PJH3 A0A2K5RE66 A0A2K6FP00
A0A194PMW9 A0A212FJV1 D6WD90 A0A1Y1M9I5 A0A336KF26 A0A336MJC5 A0A2J7PJ57 W8BUG2 A0A0A1WSX0 A0A084W143 A0A0L0CB09 A0A067QQD0 A0A1I8NT42 A0A1I8NT35 A0A034V452 N6TWN3 A0A1Q3F1K4 A0A0K8UPS4 E0VWC9 A0A1Q3F1X7 A0A182N9A6 A0A1I8M450 A0A2M4APT5 A0A1I8MVI4 A0A1A9XQZ6 W5JV16 A0A2M4ANH7 A0A1A9UP15 A0A182JFD2 A0A1B0DIE3 A0A1A9Z922 A0A182PIJ1 A0A182TEN0 A0A182WWI2 B4LG52 A0A182I6R2 Q7QIL5 A0A182L3N3 A0A0P6IVI1 A0A1B0G7J1 A0A182WBI4 A0A182MFL8 B0W5M0 A0A1B6GYE8 B4KXL0 A0A1B0C2P6 A0A0A9Z2H7 B4MM57 A0A182FEM3 A0A069DV92 A0A182JRN8 A0A182Y2B7 A0A1S4GTP2 A0A146L1M0 A0A0V0G5F8 B4IXF5 P26802 A0A1W4UGI4 A0A1A9W4X8 A0A3B0KLV4 A0A3B0KUD8 B4PKA4 B3NDI1 A0A182VAG1 A0A182RJL1 A0A224X9K7 A0A182Q8N8 B3M4E5 B4QMY0 Q29DT5 A0A023EZG8 A0A1B6C6W5 B4HK61 T1PJQ9 A0A0M5J467 A0A2K6P3N6 A0A2R2MRE2 A0A2P8ZHG2 G1M4Z3 A0A1U7R9Y5 A0A2K5UU65 A0A1J1IBZ4 A0A2K5IPK8 A0A287BGR7 A0A151N4A6 A0A091N2K9 D2HLG6 A0A2K6BYN8 A0A2K5RE81 A0A2K5M739 G7PJH3 A0A2K5RE66 A0A2K6FP00
PDB
6EM5
E-value=1.19245e-39,
Score=412
Ontologies
GO
PANTHER
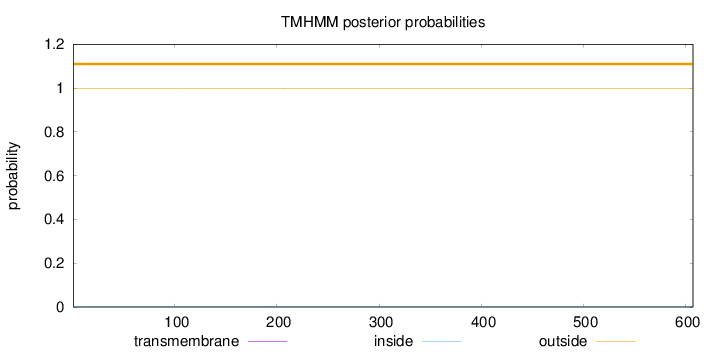
Topology
Subcellular location
Length:
607
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0168899999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00323
outside
1 - 607
Population Genetic Test Statistics
Pi
223.280975
Theta
190.314837
Tajima's D
0.695422
CLR
0.253463
CSRT
0.578321083945803
Interpretation
Uncertain
