Gene
KWMTBOMO02642
Pre Gene Modal
BGIBMGA002647
Annotation
erythrocyte_carbonic_anhydrase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.324 Mitochondrial Reliability : 1.006
Sequence
CDS
ATGGCAGACAACGAATGGGGATATACCTGTGAAAACGGTCCGGCGACATGGATAGAGAAGTTTCCTCAAGCACGTGGCACTCGACAAAGTCCCGTCGATATTGTGACTTCTCGCGTCAAAAGTGGAGCTTCCCTGCCGCCATTGAAATGGCGGTACTCTGTCAATCATCCTCGTTCCATCGTCAATCCTGGCTACTGTTGGCGTGTCGACGAAAACGGCTATGATTCAGAATTAAAAGGTGGACCTCTCAACAACGATGTTTACAAATTACAACAATGGCATTGCCACTGGGGAGCGCTCAACGGGGAGGGCTCAGAACATACAGTGGACGGTCGCTCGTTCTCCGGAGAACTCCACCTGGTTCACTGGAACACAAGCAAGTACCACAGCTTCGGGGAGGCGGCAGGAAAACCAGATGGACTAGCCGTTTTAGGAGTCCTATTAATGGTTGGCTCGAAACACCGAGAGCTCGATAAAGTTGTACAGTTACTGCCTTTCGTGCAACACAAAGGCGATAAAGTTACATTTTGCGAGCCCCTCGACCCTGCCAAGCTGCTACCAACCAAGACAGCTTACTGGACGTACCCAGGGTCTTTGACAACACCACCGTGCACGGAGTCGGTGATTTGGCTCCTCTTCAAAGAACCCGTACAAGTTTCCGCTGAACAGCTCTCTTTGATGCGGAAGTTGAAATGCGGTGAAGCGTCATGCGGCGTTGAGGCGATGGAGCTGCTCCACAATTACCGGCCCACGCTGCCCCTCGGCAACAGGGAGTTGCGCGAGTACGGCGGCAACTAG
Protein
MADNEWGYTCENGPATWIEKFPQARGTRQSPVDIVTSRVKSGASLPPLKWRYSVNHPRSIVNPGYCWRVDENGYDSELKGGPLNNDVYKLQQWHCHWGALNGEGSEHTVDGRSFSGELHLVHWNTSKYHSFGEAAGKPDGLAVLGVLLMVGSKHRELDKVVQLLPFVQHKGDKVTFCEPLDPAKLLPTKTAYWTYPGSLTTPPCTESVIWLLFKEPVQVSAEQLSLMRKLKCGEASCGVEAMELLHNYRPTLPLGNRELREYGGN
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
Q2F607
A0A212FJV0
A0A2A4K6J6
A0A2W1BIP4
A0A194RKZ8
A0A2H1VS87
+ More
H9IZG2 A0A0L7LW19 A0A194PPV8 A0A1I8NRL5 D3TM95 U5END8 T1PBP1 A0A1I8N1V6 A0A023ENB2 A0A2M4BWI9 T1DPU1 A0A2M4CU32 A0A2M4A140 A0A023EM65 Q9V396 A0A2M3YZI2 B4HX95 B4Q559 B3MN32 A0A1W4V7E5 Q3YMV3 Q17BL7 B4P3P9 A0A0A1WIM4 A0A034WJI6 B4KIX4 A0A182QUC6 A0A0K8V010 A0A1B0FDE7 B3NAD8 A0A182VKJ1 Q19MS3 A0A1B0B7M0 A0A1A9V9J8 B4N159 B4M8K9 A0A1A9XL01 A0A1L8DFB6 Q29K70 B4GWZ7 B4JCW9 A0A0M4ER24 W8AUP2 A0A182GXZ3 A0A1B0A529 A0A1A9W052 A0A1Q3G586 A0A1Q3G588 A0A1B0GN58 A0A3B0JTM5 A0A336LZX4 A0A1J1HMQ2 A0A0L0BQQ2 A0A182HR28 A0A182MMY9 A0A2A3EJM1 V9IFZ3 E1ZVE3 K7IQG1 A0A088A5W1 A0A182FN60 A0A1B6FXQ8 A0A0C9RYH4 A0A1B6DU34 A0A182PM94 A0A182R1S0 A0A084WQ40 A0A182Y642 A0A140KFJ8 A0A026W6G5 E2BC97 A0A182JWM3 A0A1B6EM51 A0A182U759 E9J6B1 A0A1B0CSY1 A0A067QN01 A0A1B6CJH3 A0A182W8K4 A0A0L7R1Z2 A0A0C9RLJ8 A0A3L8DV14 A0A182NNU3 A0A182XHA9 A0A158NBM5 A0A224XHU4 A0A023F836 B0W447 A0A0V0G702 A0A069DQP4 A0A0P4W0C2 A0A0A9X516 A0A1W4WKL6 A0A146KNY1
H9IZG2 A0A0L7LW19 A0A194PPV8 A0A1I8NRL5 D3TM95 U5END8 T1PBP1 A0A1I8N1V6 A0A023ENB2 A0A2M4BWI9 T1DPU1 A0A2M4CU32 A0A2M4A140 A0A023EM65 Q9V396 A0A2M3YZI2 B4HX95 B4Q559 B3MN32 A0A1W4V7E5 Q3YMV3 Q17BL7 B4P3P9 A0A0A1WIM4 A0A034WJI6 B4KIX4 A0A182QUC6 A0A0K8V010 A0A1B0FDE7 B3NAD8 A0A182VKJ1 Q19MS3 A0A1B0B7M0 A0A1A9V9J8 B4N159 B4M8K9 A0A1A9XL01 A0A1L8DFB6 Q29K70 B4GWZ7 B4JCW9 A0A0M4ER24 W8AUP2 A0A182GXZ3 A0A1B0A529 A0A1A9W052 A0A1Q3G586 A0A1Q3G588 A0A1B0GN58 A0A3B0JTM5 A0A336LZX4 A0A1J1HMQ2 A0A0L0BQQ2 A0A182HR28 A0A182MMY9 A0A2A3EJM1 V9IFZ3 E1ZVE3 K7IQG1 A0A088A5W1 A0A182FN60 A0A1B6FXQ8 A0A0C9RYH4 A0A1B6DU34 A0A182PM94 A0A182R1S0 A0A084WQ40 A0A182Y642 A0A140KFJ8 A0A026W6G5 E2BC97 A0A182JWM3 A0A1B6EM51 A0A182U759 E9J6B1 A0A1B0CSY1 A0A067QN01 A0A1B6CJH3 A0A182W8K4 A0A0L7R1Z2 A0A0C9RLJ8 A0A3L8DV14 A0A182NNU3 A0A182XHA9 A0A158NBM5 A0A224XHU4 A0A023F836 B0W447 A0A0V0G702 A0A069DQP4 A0A0P4W0C2 A0A0A9X516 A0A1W4WKL6 A0A146KNY1
Pubmed
22118469
28756777
26354079
19121390
26227816
20353571
+ More
25315136 24945155 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17510324 17550304 25830018 25348373 17981859 15632085 24495485 26483478 26108605 20798317 20075255 24438588 25244985 24508170 21282665 24845553 30249741 21347285 25474469 26334808 27129103 25401762 26823975
25315136 24945155 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17510324 17550304 25830018 25348373 17981859 15632085 24495485 26483478 26108605 20798317 20075255 24438588 25244985 24508170 21282665 24845553 30249741 21347285 25474469 26334808 27129103 25401762 26823975
EMBL
DQ311265
ABD36210.1
AGBW02008206
OWR54018.1
NWSH01000078
PCG79861.1
+ More
KZ150098 PZC73575.1 KQ460207 KPJ16651.1 ODYU01004121 SOQ43658.1 BABH01014126 JTDY01000024 KOB79366.1 KQ459597 KPI94769.1 EZ422547 ADD18823.1 GANO01000635 JAB59236.1 KA646182 AFP60811.1 GAPW01003210 GAPW01003209 GAPW01003208 JAC10390.1 GGFJ01008233 MBW57374.1 GAMD01003039 JAA98551.1 GGFL01004597 MBW68775.1 GGFK01001148 MBW34469.1 GAPW01003211 JAC10387.1 AE014134 AY058327 DQ062772 AAF53332.1 AAL13556.1 AAY56645.1 GGFM01000929 MBW21680.1 CH480818 EDW51675.1 CM000361 CM002910 EDX04965.1 KMY90158.1 CH902620 EDV32010.1 DQ062773 AAY56646.1 CH477320 EAT43649.1 CM000157 EDW88350.1 GBXI01016059 JAC98232.1 GAKP01005000 GAKP01004999 GAKP01004998 JAC53953.1 CH933807 EDW13487.1 AXCN02001986 GDHF01022788 GDHF01020090 GDHF01003279 JAI29526.1 JAI32224.1 JAI49035.1 CCAG010008796 CCAG010008797 CCAG010008798 CH954177 EDV58640.1 DQ518576 ABF66618.1 JXJN01009688 CH963920 EDW78040.1 CH940654 EDW57535.1 GFDF01008932 JAV05152.1 CH379061 EAL33308.1 CH479195 EDW27324.1 CH916368 EDW03208.1 CP012523 ALC39550.1 GAMC01016618 JAB89937.1 JXUM01020699 JXUM01020700 GFDL01000074 JAV34971.1 GFDL01000073 JAV34972.1 AJVK01004379 OUUW01000010 SPP85455.1 UFQS01000363 UFQT01000363 SSX03176.1 SSX23542.1 CVRI01000010 CRK88802.1 JRES01001517 KNC22323.1 APCN01001635 AXCM01001635 KZ288229 PBC31674.1 JR041354 AEY59421.1 GL434492 EFN74853.1 GECZ01014792 JAS54977.1 GBYB01013021 JAG82788.1 GEDC01008114 JAS29184.1 ATLV01025177 KE525371 KFB52334.1 LC126080 BAU68125.1 KK107372 EZA51697.1 GL447257 EFN86633.1 GECZ01030791 JAS38978.1 GL768230 EFZ11655.1 AJWK01026760 AJWK01026761 AJWK01026762 KK853231 KDR09625.1 GEDC01023664 JAS13634.1 KQ414667 KOC64838.1 GBYB01014232 JAG83999.1 QOIP01000004 RLU23739.1 ADTU01011224 ADTU01011225 ADTU01011226 GFTR01004371 JAW12055.1 GBBI01001287 JAC17425.1 DS231834 EDS32640.1 GECL01002387 JAP03737.1 GBGD01002536 JAC86353.1 GDKW01000288 JAI56307.1 GBHO01029697 GBHO01029696 JAG13907.1 JAG13908.1 GDHC01021234 JAP97394.1
KZ150098 PZC73575.1 KQ460207 KPJ16651.1 ODYU01004121 SOQ43658.1 BABH01014126 JTDY01000024 KOB79366.1 KQ459597 KPI94769.1 EZ422547 ADD18823.1 GANO01000635 JAB59236.1 KA646182 AFP60811.1 GAPW01003210 GAPW01003209 GAPW01003208 JAC10390.1 GGFJ01008233 MBW57374.1 GAMD01003039 JAA98551.1 GGFL01004597 MBW68775.1 GGFK01001148 MBW34469.1 GAPW01003211 JAC10387.1 AE014134 AY058327 DQ062772 AAF53332.1 AAL13556.1 AAY56645.1 GGFM01000929 MBW21680.1 CH480818 EDW51675.1 CM000361 CM002910 EDX04965.1 KMY90158.1 CH902620 EDV32010.1 DQ062773 AAY56646.1 CH477320 EAT43649.1 CM000157 EDW88350.1 GBXI01016059 JAC98232.1 GAKP01005000 GAKP01004999 GAKP01004998 JAC53953.1 CH933807 EDW13487.1 AXCN02001986 GDHF01022788 GDHF01020090 GDHF01003279 JAI29526.1 JAI32224.1 JAI49035.1 CCAG010008796 CCAG010008797 CCAG010008798 CH954177 EDV58640.1 DQ518576 ABF66618.1 JXJN01009688 CH963920 EDW78040.1 CH940654 EDW57535.1 GFDF01008932 JAV05152.1 CH379061 EAL33308.1 CH479195 EDW27324.1 CH916368 EDW03208.1 CP012523 ALC39550.1 GAMC01016618 JAB89937.1 JXUM01020699 JXUM01020700 GFDL01000074 JAV34971.1 GFDL01000073 JAV34972.1 AJVK01004379 OUUW01000010 SPP85455.1 UFQS01000363 UFQT01000363 SSX03176.1 SSX23542.1 CVRI01000010 CRK88802.1 JRES01001517 KNC22323.1 APCN01001635 AXCM01001635 KZ288229 PBC31674.1 JR041354 AEY59421.1 GL434492 EFN74853.1 GECZ01014792 JAS54977.1 GBYB01013021 JAG82788.1 GEDC01008114 JAS29184.1 ATLV01025177 KE525371 KFB52334.1 LC126080 BAU68125.1 KK107372 EZA51697.1 GL447257 EFN86633.1 GECZ01030791 JAS38978.1 GL768230 EFZ11655.1 AJWK01026760 AJWK01026761 AJWK01026762 KK853231 KDR09625.1 GEDC01023664 JAS13634.1 KQ414667 KOC64838.1 GBYB01014232 JAG83999.1 QOIP01000004 RLU23739.1 ADTU01011224 ADTU01011225 ADTU01011226 GFTR01004371 JAW12055.1 GBBI01001287 JAC17425.1 DS231834 EDS32640.1 GECL01002387 JAP03737.1 GBGD01002536 JAC86353.1 GDKW01000288 JAI56307.1 GBHO01029697 GBHO01029696 JAG13907.1 JAG13908.1 GDHC01021234 JAP97394.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000005204
UP000037510
UP000053268
+ More
UP000095300 UP000095301 UP000000803 UP000001292 UP000000304 UP000007801 UP000192221 UP000008820 UP000002282 UP000009192 UP000075886 UP000092444 UP000008711 UP000075903 UP000092460 UP000078200 UP000007798 UP000008792 UP000092443 UP000001819 UP000008744 UP000001070 UP000092553 UP000069940 UP000092445 UP000091820 UP000092462 UP000268350 UP000183832 UP000037069 UP000075840 UP000075883 UP000242457 UP000000311 UP000002358 UP000005203 UP000069272 UP000075885 UP000075900 UP000030765 UP000076408 UP000053097 UP000008237 UP000075881 UP000075902 UP000092461 UP000027135 UP000075920 UP000053825 UP000279307 UP000075884 UP000076407 UP000005205 UP000002320 UP000192223
UP000095300 UP000095301 UP000000803 UP000001292 UP000000304 UP000007801 UP000192221 UP000008820 UP000002282 UP000009192 UP000075886 UP000092444 UP000008711 UP000075903 UP000092460 UP000078200 UP000007798 UP000008792 UP000092443 UP000001819 UP000008744 UP000001070 UP000092553 UP000069940 UP000092445 UP000091820 UP000092462 UP000268350 UP000183832 UP000037069 UP000075840 UP000075883 UP000242457 UP000000311 UP000002358 UP000005203 UP000069272 UP000075885 UP000075900 UP000030765 UP000076408 UP000053097 UP000008237 UP000075881 UP000075902 UP000092461 UP000027135 UP000075920 UP000053825 UP000279307 UP000075884 UP000076407 UP000005205 UP000002320 UP000192223
Pfam
PF00194 Carb_anhydrase
Interpro
Gene 3D
ProteinModelPortal
Q2F607
A0A212FJV0
A0A2A4K6J6
A0A2W1BIP4
A0A194RKZ8
A0A2H1VS87
+ More
H9IZG2 A0A0L7LW19 A0A194PPV8 A0A1I8NRL5 D3TM95 U5END8 T1PBP1 A0A1I8N1V6 A0A023ENB2 A0A2M4BWI9 T1DPU1 A0A2M4CU32 A0A2M4A140 A0A023EM65 Q9V396 A0A2M3YZI2 B4HX95 B4Q559 B3MN32 A0A1W4V7E5 Q3YMV3 Q17BL7 B4P3P9 A0A0A1WIM4 A0A034WJI6 B4KIX4 A0A182QUC6 A0A0K8V010 A0A1B0FDE7 B3NAD8 A0A182VKJ1 Q19MS3 A0A1B0B7M0 A0A1A9V9J8 B4N159 B4M8K9 A0A1A9XL01 A0A1L8DFB6 Q29K70 B4GWZ7 B4JCW9 A0A0M4ER24 W8AUP2 A0A182GXZ3 A0A1B0A529 A0A1A9W052 A0A1Q3G586 A0A1Q3G588 A0A1B0GN58 A0A3B0JTM5 A0A336LZX4 A0A1J1HMQ2 A0A0L0BQQ2 A0A182HR28 A0A182MMY9 A0A2A3EJM1 V9IFZ3 E1ZVE3 K7IQG1 A0A088A5W1 A0A182FN60 A0A1B6FXQ8 A0A0C9RYH4 A0A1B6DU34 A0A182PM94 A0A182R1S0 A0A084WQ40 A0A182Y642 A0A140KFJ8 A0A026W6G5 E2BC97 A0A182JWM3 A0A1B6EM51 A0A182U759 E9J6B1 A0A1B0CSY1 A0A067QN01 A0A1B6CJH3 A0A182W8K4 A0A0L7R1Z2 A0A0C9RLJ8 A0A3L8DV14 A0A182NNU3 A0A182XHA9 A0A158NBM5 A0A224XHU4 A0A023F836 B0W447 A0A0V0G702 A0A069DQP4 A0A0P4W0C2 A0A0A9X516 A0A1W4WKL6 A0A146KNY1
H9IZG2 A0A0L7LW19 A0A194PPV8 A0A1I8NRL5 D3TM95 U5END8 T1PBP1 A0A1I8N1V6 A0A023ENB2 A0A2M4BWI9 T1DPU1 A0A2M4CU32 A0A2M4A140 A0A023EM65 Q9V396 A0A2M3YZI2 B4HX95 B4Q559 B3MN32 A0A1W4V7E5 Q3YMV3 Q17BL7 B4P3P9 A0A0A1WIM4 A0A034WJI6 B4KIX4 A0A182QUC6 A0A0K8V010 A0A1B0FDE7 B3NAD8 A0A182VKJ1 Q19MS3 A0A1B0B7M0 A0A1A9V9J8 B4N159 B4M8K9 A0A1A9XL01 A0A1L8DFB6 Q29K70 B4GWZ7 B4JCW9 A0A0M4ER24 W8AUP2 A0A182GXZ3 A0A1B0A529 A0A1A9W052 A0A1Q3G586 A0A1Q3G588 A0A1B0GN58 A0A3B0JTM5 A0A336LZX4 A0A1J1HMQ2 A0A0L0BQQ2 A0A182HR28 A0A182MMY9 A0A2A3EJM1 V9IFZ3 E1ZVE3 K7IQG1 A0A088A5W1 A0A182FN60 A0A1B6FXQ8 A0A0C9RYH4 A0A1B6DU34 A0A182PM94 A0A182R1S0 A0A084WQ40 A0A182Y642 A0A140KFJ8 A0A026W6G5 E2BC97 A0A182JWM3 A0A1B6EM51 A0A182U759 E9J6B1 A0A1B0CSY1 A0A067QN01 A0A1B6CJH3 A0A182W8K4 A0A0L7R1Z2 A0A0C9RLJ8 A0A3L8DV14 A0A182NNU3 A0A182XHA9 A0A158NBM5 A0A224XHU4 A0A023F836 B0W447 A0A0V0G702 A0A069DQP4 A0A0P4W0C2 A0A0A9X516 A0A1W4WKL6 A0A146KNY1
PDB
4HBA
E-value=1.28241e-52,
Score=520
Ontologies
PATHWAY
GO
PANTHER
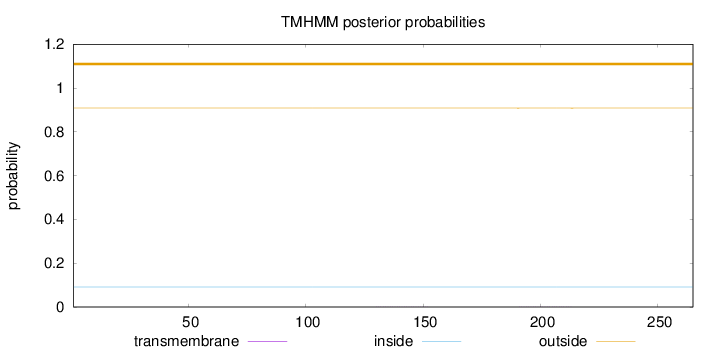
Topology
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01845
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09129
outside
1 - 265
Population Genetic Test Statistics
Pi
267.407808
Theta
193.369025
Tajima's D
1.040261
CLR
0.289689
CSRT
0.668016599170042
Interpretation
Uncertain
