Gene
KWMTBOMO02641
Annotation
PREDICTED:_uncharacterized_protein_LOC101740474_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.085 Extracellular Reliability : 1.006 Nuclear Reliability : 1.567
Sequence
CDS
ATGGTGTTAAATATTTGTGCTGTGTACATCCCATCTCCTTTGAATGTTGATTATGTCGACATTTTTACCAAAAATACTGTGCAGGTTATTCACAACAACATGACAAAAGGTATTACTGAACCATTATTTCTTTTAGTAGGAGACTTTAATATGGGGGATATTGAGTGGTGTAAAGATAATTCTTCTGCATATATGACCCCATCAACATCAGGGAATAATGTCTATACAGAATTTTTAGATGCCATGTCTTACTGCGACCTTTATCAGTTTAATGATATGATCAACGCTAATAATCGAATCCTTGACTTGGTATTTAGTGGTGCGTCAAGTCATATCTCAATTGAATCTGCCAAGGGCTTTTCGACTGCACCTGACCCATACCATCCGCCACTGGACATTGCATTTCAAGAAACATCTGTGCATGTTGTGAAACCTGACTCTGCCCCAGCATTAAACTACTTCAAATCCAATTATGAGGCTCTGAATAAAGACTTAAAGCAGGTTGATTGGACCAACTTACTACAAGAATCATATTCTGTGGATACAATGAATGCTCCCAATCATCTCAAAAACCGTCCGAAATACTTCTGGAAAATTTTTAAGGATAAAACAGCTAATAGTCGTTCTTTGCCTAACGAAATGAAGCTTGGTCGGGTAGAGGCTAAGGGAGGACAGGAGATCTCTGAATTATTCTCTCAACATTTCCAAAGTGTGTATGATTCCAACATTAATCATGAAACAAGTTGCTCCTCTCATTATTGTAATGTGTCCGGCAATGCATCATATAATATCGTTGAAGTTAATGAAACCGAAGTTTTAAATCAGCTAAAGCTTTTAAATGCGGACAAAGGACCTGGCCCTGATTTGATACCTCCTATATTCATAACAAAATGTGCTCTAAGTTTATATAAACCTCTAGCCAAAATTTATACCAAATCTCTCCAAAGTGGTGCTTACCCCACTCAGTGGAAATTAGCGCACTTAACACCGATCCACAAGGATGGGAGGCTGGATGATATAGCCCAATATCGACCTGTCTCCATACTGTCTGGGTTTGGCAAGGTATTGGAAGCCATAGTTAATAAAAAACTGATAACTCATTTCAAGCAATCTATAGACACAAATCAACACGGCTTTCTGCCATCCAAATCGACAAATACTAACTTAGTTAGTTATGTTTCTGATATAATTGATACTATGGACGGAGGTGGCGAGGTGCACTCTATCTACACCGATTTCAGAAAGGCGTTTGACCTAGTTAACCATAACTGCCTATTACACAAGCTTGCAGAACTGGGCATCCATGGATCGTTACTGAGATGGTGCGAGTCGTACATACGCAACCGCTCGCAATTGGTAAACATTCAAGGGTTTAAATCTAGAGTGCACATGGTCCCATCAGGGGTCCCTCAAGGGTCTCATCTAGGTCCTTTCTTTTTCCTAGTCTACATCAATAACATTAAGCATGTCATCCAATCTAAATTTAAATTATATGCTGATGACTTAAAAGTGTATCGTGCTGTTGAAGACCATTCGGATATAATGGCTCTTCAGTCAGATATCAATGCGATTTACAAATGGAGTACGCAGAATTACATGCACTTGAATTTGCAAAAGAAAAATAATCAAGACCATGATGAGGCTGAAGATGAAATCATGACCAATTAA
Protein
MVLNICAVYIPSPLNVDYVDIFTKNTVQVIHNNMTKGITEPLFLLVGDFNMGDIEWCKDNSSAYMTPSTSGNNVYTEFLDAMSYCDLYQFNDMINANNRILDLVFSGASSHISIESAKGFSTAPDPYHPPLDIAFQETSVHVVKPDSAPALNYFKSNYEALNKDLKQVDWTNLLQESYSVDTMNAPNHLKNRPKYFWKIFKDKTANSRSLPNEMKLGRVEAKGGQEISELFSQHFQSVYDSNINHETSCSSHYCNVSGNASYNIVEVNETEVLNQLKLLNADKGPGPDLIPPIFITKCALSLYKPLAKIYTKSLQSGAYPTQWKLAHLTPIHKDGRLDDIAQYRPVSILSGFGKVLEAIVNKKLITHFKQSIDTNQHGFLPSKSTNTNLVSYVSDIIDTMDGGGEVHSIYTDFRKAFDLVNHNCLLHKLAELGIHGSLLRWCESYIRNRSQLVNIQGFKSRVHMVPSGVPQGSHLGPFFFLVYINNIKHVIQSKFKLYADDLKVYRAVEDHSDIMALQSDINAIYKWSTQNYMHLNLQKKNNQDHDEAEDEIMTN
Summary
Uniprot
EMBL
ABLF02022753
ABLF02028025
ABLF02028028
UZAG01001120
VDO10445.1
GGMR01012775
+ More
MBY25394.1 GGMR01013083 MBY25702.1 GEZM01075215 JAV64203.1 GEHC01000951 JAV46694.1 ABLF02041910 GEHC01000982 JAV46663.1 ABLF02027115 ABLF02027120 ABLF02055498 ABLF02012254 ABLF02012256 ABLF02012260 ABLF02041969 GGMR01010742 MBY23361.1 GGMR01014615 MBY27234.1 NWSH01000614 PCG75292.1 GGMS01015155 MBY84358.1 GGMR01016676 MBY29295.1 ABLF02041544 GDUN01000801 JAN95118.1 GEFM01006330 JAP69466.1 GDUN01000797 JAN95122.1
MBY25394.1 GGMR01013083 MBY25702.1 GEZM01075215 JAV64203.1 GEHC01000951 JAV46694.1 ABLF02041910 GEHC01000982 JAV46663.1 ABLF02027115 ABLF02027120 ABLF02055498 ABLF02012254 ABLF02012256 ABLF02012260 ABLF02041969 GGMR01010742 MBY23361.1 GGMR01014615 MBY27234.1 NWSH01000614 PCG75292.1 GGMS01015155 MBY84358.1 GGMR01016676 MBY29295.1 ABLF02041544 GDUN01000801 JAN95118.1 GEFM01006330 JAP69466.1 GDUN01000797 JAN95122.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
PDB
5G2X
E-value=0.0338566,
Score=88
Ontologies
GO
Topology
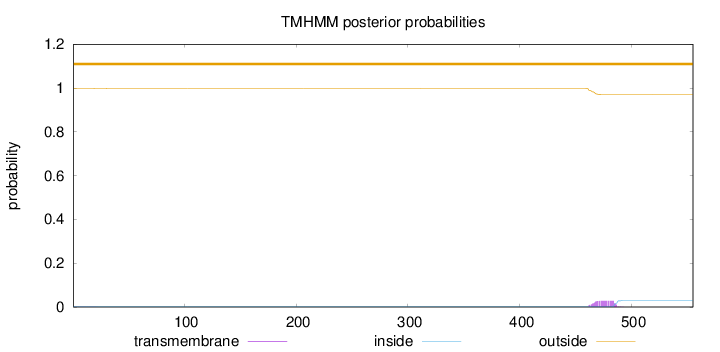
Length:
555
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.61413
Exp number, first 60 AAs:
0.00631999999999999
Total prob of N-in:
0.00156
outside
1 - 555
Population Genetic Test Statistics
Pi
203.165829
Theta
186.182357
Tajima's D
1.348097
CLR
235.439681
CSRT
0.749862506874656
Interpretation
Uncertain
