Gene
KWMTBOMO02640
Pre Gene Modal
BGIBMGA002645
Annotation
PREDICTED:_A_disintegrin_and_metalloproteinase_with_thrombospondin_motifs_14-like_[Bombyx_mori]
Full name
Histone-lysine N-methyltransferase, H3 lysine-79 specific
Alternative Name
Histone H3-K79 methyltransferase
Location in the cell
Extracellular Reliability : 1.851 Nuclear Reliability : 1.308
Sequence
CDS
ATGGCGTATCGAAATTTCCAGGAGCTTCTGCCAACATTTGGGTGGTGGCAGGAGGAGGGGCGTGGTGAGCGGGGTCACGGCGCAGACGTACAAGTAGTGTACTTGCCAGCGTTGTTGCCGCGGGAAGCACAGTCCTCCGTGGACGGTAGCAATGATGACGTTCCTTTACCGTACAACTTTGATGCGTTCGGTCGTAACTTCGATCTTCAGCTGGTGCCAAACCGTCGACTGGTGTCTCCAGAGTTCCGCGTGTGGACCGAACGTGGGGAAGAGAAACCCTTGTCCAGCGTGGATGCCTCTTGCCATTTCCTGCATGCTAGTCCTAACGCAGTAGCTGCCTTCTCGGCTTGCAAAGATCATGCACTACACGGACTAATCGTCGTCGATAACTCTACATATGAGGTCCGCCCGTTGCCTTCTGGTGTTGGCCGTGCTGAGAACAGCTATGGTCGTACTGCACACGTAATTCGAAGAGCATCTCCTCCTTCACCGCCCGTCGATGATGCCAGAGTCCTAAGACCGCGGCCGCGGCGCCGTCCGCGAAACCACACTCCTCCAGTGAAGAAAGGCCCGGCCAGCTATACCATAGAAATAGCACTGTTTCTCGACGAAGCTGCATATAAAATTTTTCATCCATTTTTTCATTACAATGAAGCTGATTTGCGTGATATGTTGCTGGCTTATATAAATGGGGTACAGGCGCTATATCAACATTCTTCCCTTGGTACTCATATCCAGTTGTCACTAGTGAGGTTAACGTTATTAAGGTCTCAGCCAACTGATCTTTCAGCGCATGCCGAACGTGGCAAACTCCTAGACTCTTTCTGTGCATATCAAAGATCACTAAACGTTGAAGATGATGAAGATCCCGAACATTGGGATATGGCCTTACTGTTATCTGGATTAGATTTTTATTCAGAAGAAAATGGTCGGCGGAACGGCGTCACTATGGGCCTAGCGCCCGTGGGTGGGGTGTGCTTGCCGGCGCACGCATGCGTGGTGTCGGAGTTCGGTGCCACGGACGTGCTCGGCCGTCCCTACCCGTCAGCAGGCTTCACTTCCGTCTACATACTGGCGCACGAGATCGGACACAACTTGGGTATGCATCACGACGGTACCGGGAACACGTGCGCCCGCGACGGTTACATAATGTCGCCGTCTCGAGGAACCAATGGAGAGGCTTCCTGGTCATCCTGCAGTGCTAGAGTCGTTGCCGACCTCAAATGGGCTACTTGCCTGTTGGATGGTGACGACGGCGAGGAAGACATCCCAAATGAGCTTCAGCATGAACGATTTAGCGGAGCGCCTGGTGCTATATGGAGCGCAAAGAAACAGTGTGAGGTTTTGTTGCGTGACGCGGACGCAGCGCCTTGGTCGGGCGGCGGAGACCAGCGGGGGCAATGCGCCCAGCTCGCGTGCCGCACCCCGCACCGCGCCGGCTTCTTCTACGCCGGCCCCGCGCTGCCCGGCACTGCTTGTGGCAATAAAATGTGGTGTCAAGGTGGGGAGTGTGTGCCTTCGAACTCCGTACCGACATCCAGCACGCGCGTTCCGCCATCGGCCGGTATGGCGGAGGGCGAGTCTGGCGGCTGGACCGTATGGAGAGAAGGCGCGTGTCGGTCCGGCTGCTCAGCCAAGTCGCGGGGCTTTAGAGAACGTAGACGGGAATGCGCTACGACCACCACCTGCCAAGGATCCGCTTACGATGTCGTACTGTGTGATGATACCAAGGTTTGCGGCAAGAAGAGCCGAGTGAGTGCCAATGAGCTTGCTTCCCGCAAGTGCAGCGAGTACGCGGCGCGCGTGCCGTCGCTGGACGCGCGCGCCGGCGGGCTGCAGGCCCCGCATGATCCTACTCGCATGTGGATGGGGTGCGCAATATTCTGCAGGCGAGCTTCCGGCGGTGGGTTTTACGCTCCTCGGGTGGAACTCAACGCGGCAGGTCTCGATCCCTATCTGCCTGACGGTACCTGGTGCCACAACGATGGGAAACATGACTATTACTGCATGCAACATCATTGTCTGCCAGAGAATTTCAAGATGTCAACGCAATACCACATTTGGGAGTTGCCATCTCAGGACATTGGCGGGTCGTTTAACGCTCGTGCTTTGACCACTCCTGATGATGAAGCCGCCGAGGCGGCTGTACGAGAATATTTATCTTTAGACAGCCATGGAGTGCCTCTTACGAGAGGCGCATTCCCTCCAAGTTTACCCGATGAACCTGAAGACAACTGGGAGGTAGTCGACTACGTGGAAATTCATAGAAATGACCCCTCTTAA
Protein
MAYRNFQELLPTFGWWQEEGRGERGHGADVQVVYLPALLPREAQSSVDGSNDDVPLPYNFDAFGRNFDLQLVPNRRLVSPEFRVWTERGEEKPLSSVDASCHFLHASPNAVAAFSACKDHALHGLIVVDNSTYEVRPLPSGVGRAENSYGRTAHVIRRASPPSPPVDDARVLRPRPRRRPRNHTPPVKKGPASYTIEIALFLDEAAYKIFHPFFHYNEADLRDMLLAYINGVQALYQHSSLGTHIQLSLVRLTLLRSQPTDLSAHAERGKLLDSFCAYQRSLNVEDDEDPEHWDMALLLSGLDFYSEENGRRNGVTMGLAPVGGVCLPAHACVVSEFGATDVLGRPYPSAGFTSVYILAHEIGHNLGMHHDGTGNTCARDGYIMSPSRGTNGEASWSSCSARVVADLKWATCLLDGDDGEEDIPNELQHERFSGAPGAIWSAKKQCEVLLRDADAAPWSGGGDQRGQCAQLACRTPHRAGFFYAGPALPGTACGNKMWCQGGECVPSNSVPTSSTRVPPSAGMAEGESGGWTVWREGACRSGCSAKSRGFRERRRECATTTTCQGSAYDVVLCDDTKVCGKKSRVSANELASRKCSEYAARVPSLDARAGGLQAPHDPTRMWMGCAIFCRRASGGGFYAPRVELNAAGLDPYLPDGTWCHNDGKHDYYCMQHHCLPENFKMSTQYHIWELPSQDIGGSFNARALTTPDDEAAEAAVREYLSLDSHGVPLTRGAFPPSLPDEPEDNWEVVDYVEIHRNDPS
Summary
Catalytic Activity
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Miscellaneous
In contrast to other lysine histone methyltransferases, it does not contain a SET domain, suggesting the existence of another mechanism for methylation of lysine residues of histones.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. DOT1 family.
Uniprot
H9IZG0
A0A2A4K869
A0A2W1BL44
A0A2H1VZM7
A0A212FJY1
A0A194PMV9
+ More
A0A194RH49 A0A0L7LV37 D6WRL2 A0A2J7RCF5 A0A2A3EIQ2 A0A0C9QT06 A0A088A5V6 A0A0M8ZTR7 A0A0L7R1T7 A0A1B0EVJ6 A0A195BAH8 A0A158NUV5 A0A151K0J4 E2A767 F4WE17 A0A3L8D6J8 U4UQG6 A0A151IV55 A0A026WD85 A0A151I6G1 B0W448 A0A232F4Y4 A0A151WP70 A0A182W8K3 A0A084WQ39 A0A182PM95 J9HSG8 A0A182XHA8 A0A1J1ISI4 A0A182R1R9 A0A182HR27 A0A182KH83 A0A182FN59 A0A182Q2D9 A0A154P3M5 A0A182NNU2 K7JNF7 A0A2S2NN49 N6U222 J9JKB5 E0VIC6 A0A182GXZ2 A0A2S2Q9Q2 A0A0N8CFH1 A0A0P5ND54 A0A0N8B5Q9 A0A0P5ZV18 A0A164NEH7 A0A0P5XRH6 A0A0P5XLF5 A0A0N8C3N1 A0A0N8B670 E9GBH0 A0A0P5D8W7 A0A0P6GPU8 A0A0P5ZV22 A0A0P6IMI2 A0A0P5SGW4 A0A0P5ZVI0 A0A0P5D981 A0A0P5K7C4 A0A0N8A4C8 A0A0N8DJ19 A0A0P6HRV6 A0A0P5LTZ7 A0A0P5RDW6 A0A0N8DHU4 A0A182M5L6 A0A0P4Z4M5 A0A0P5B1N8 A0A0N8AGF6 A0A0P6CYJ7 A0A087SYI7 A0A182Y641 A0A164NDJ1 A0A0K2TDM0
A0A194RH49 A0A0L7LV37 D6WRL2 A0A2J7RCF5 A0A2A3EIQ2 A0A0C9QT06 A0A088A5V6 A0A0M8ZTR7 A0A0L7R1T7 A0A1B0EVJ6 A0A195BAH8 A0A158NUV5 A0A151K0J4 E2A767 F4WE17 A0A3L8D6J8 U4UQG6 A0A151IV55 A0A026WD85 A0A151I6G1 B0W448 A0A232F4Y4 A0A151WP70 A0A182W8K3 A0A084WQ39 A0A182PM95 J9HSG8 A0A182XHA8 A0A1J1ISI4 A0A182R1R9 A0A182HR27 A0A182KH83 A0A182FN59 A0A182Q2D9 A0A154P3M5 A0A182NNU2 K7JNF7 A0A2S2NN49 N6U222 J9JKB5 E0VIC6 A0A182GXZ2 A0A2S2Q9Q2 A0A0N8CFH1 A0A0P5ND54 A0A0N8B5Q9 A0A0P5ZV18 A0A164NEH7 A0A0P5XRH6 A0A0P5XLF5 A0A0N8C3N1 A0A0N8B670 E9GBH0 A0A0P5D8W7 A0A0P6GPU8 A0A0P5ZV22 A0A0P6IMI2 A0A0P5SGW4 A0A0P5ZVI0 A0A0P5D981 A0A0P5K7C4 A0A0N8A4C8 A0A0N8DJ19 A0A0P6HRV6 A0A0P5LTZ7 A0A0P5RDW6 A0A0N8DHU4 A0A182M5L6 A0A0P4Z4M5 A0A0P5B1N8 A0A0N8AGF6 A0A0P6CYJ7 A0A087SYI7 A0A182Y641 A0A164NDJ1 A0A0K2TDM0
EC Number
2.1.1.43
Pubmed
EMBL
BABH01014132
NWSH01000078
PCG79860.1
KZ150098
PZC73576.1
ODYU01005438
+ More
SOQ46289.1 AGBW02008206 OWR54019.1 KQ459597 KPI94766.1 KQ460207 KPJ16650.1 JTDY01000024 KOB79368.1 KQ971354 EFA07053.2 NEVH01005885 PNF38508.1 KZ288229 PBC31683.1 GBYB01006864 JAG76631.1 KQ435848 KOX70980.1 KQ414667 KOC64827.1 AJWK01020279 KQ976532 KYM81548.1 ADTU01026691 ADTU01026692 KQ981296 KYN43095.1 GL437267 EFN70723.1 GL888102 EGI67455.1 QOIP01000012 RLU15801.1 KB632319 ERL92285.1 KQ980926 KYN11392.1 KK107261 EZA53928.1 KQ978473 KYM93694.1 DS231834 EDS32641.1 NNAY01000969 OXU25672.1 KQ982905 KYQ49495.1 ATLV01025174 ATLV01025175 KE525371 KFB52333.1 CH477320 EJY57558.1 CVRI01000057 CRL02070.1 APCN01001634 AXCN02001984 KQ434809 KZC06516.1 GGMR01005994 MBY18613.1 APGK01045628 APGK01045629 APGK01045630 KB741037 ENN74671.1 ABLF02025829 ABLF02025830 DS235191 EEB13132.1 JXUM01020696 JXUM01020697 KQ560579 KXJ81822.1 GGMS01005265 MBY74468.1 GDIP01136879 JAL66835.1 GDIQ01166508 JAK85217.1 GDIQ01210187 JAK41538.1 GDIP01038649 JAM65066.1 LRGB01002860 KZS05887.1 GDIP01070392 JAM33323.1 GDIP01070393 JAM33322.1 GDIQ01118011 JAL33715.1 GDIQ01208899 JAK42826.1 GL732538 EFX83156.1 GDIP01159285 JAJ64117.1 GDIQ01030662 JAN64075.1 GDIP01039101 JAM64614.1 GDIQ01019418 JAN75319.1 GDIP01139870 JAL63844.1 GDIP01051437 JAM52278.1 GDIP01161082 JAJ62320.1 GDIQ01188393 JAK63332.1 GDIP01183713 JAJ39689.1 GDIP01028895 JAM74820.1 GDIQ01015899 JAN78838.1 GDIQ01165615 JAK86110.1 GDIQ01102775 JAL48951.1 GDIP01032295 JAM71420.1 AXCM01000626 GDIP01218616 JAJ04786.1 GDIP01191128 JAJ32274.1 GDIP01149889 JAJ73513.1 GDIQ01087935 JAN06802.1 KK112544 KFM57926.1 KZS05860.1 HACA01006752 CDW24113.1
SOQ46289.1 AGBW02008206 OWR54019.1 KQ459597 KPI94766.1 KQ460207 KPJ16650.1 JTDY01000024 KOB79368.1 KQ971354 EFA07053.2 NEVH01005885 PNF38508.1 KZ288229 PBC31683.1 GBYB01006864 JAG76631.1 KQ435848 KOX70980.1 KQ414667 KOC64827.1 AJWK01020279 KQ976532 KYM81548.1 ADTU01026691 ADTU01026692 KQ981296 KYN43095.1 GL437267 EFN70723.1 GL888102 EGI67455.1 QOIP01000012 RLU15801.1 KB632319 ERL92285.1 KQ980926 KYN11392.1 KK107261 EZA53928.1 KQ978473 KYM93694.1 DS231834 EDS32641.1 NNAY01000969 OXU25672.1 KQ982905 KYQ49495.1 ATLV01025174 ATLV01025175 KE525371 KFB52333.1 CH477320 EJY57558.1 CVRI01000057 CRL02070.1 APCN01001634 AXCN02001984 KQ434809 KZC06516.1 GGMR01005994 MBY18613.1 APGK01045628 APGK01045629 APGK01045630 KB741037 ENN74671.1 ABLF02025829 ABLF02025830 DS235191 EEB13132.1 JXUM01020696 JXUM01020697 KQ560579 KXJ81822.1 GGMS01005265 MBY74468.1 GDIP01136879 JAL66835.1 GDIQ01166508 JAK85217.1 GDIQ01210187 JAK41538.1 GDIP01038649 JAM65066.1 LRGB01002860 KZS05887.1 GDIP01070392 JAM33323.1 GDIP01070393 JAM33322.1 GDIQ01118011 JAL33715.1 GDIQ01208899 JAK42826.1 GL732538 EFX83156.1 GDIP01159285 JAJ64117.1 GDIQ01030662 JAN64075.1 GDIP01039101 JAM64614.1 GDIQ01019418 JAN75319.1 GDIP01139870 JAL63844.1 GDIP01051437 JAM52278.1 GDIP01161082 JAJ62320.1 GDIQ01188393 JAK63332.1 GDIP01183713 JAJ39689.1 GDIP01028895 JAM74820.1 GDIQ01015899 JAN78838.1 GDIQ01165615 JAK86110.1 GDIQ01102775 JAL48951.1 GDIP01032295 JAM71420.1 AXCM01000626 GDIP01218616 JAJ04786.1 GDIP01191128 JAJ32274.1 GDIP01149889 JAJ73513.1 GDIQ01087935 JAN06802.1 KK112544 KFM57926.1 KZS05860.1 HACA01006752 CDW24113.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000007266 UP000235965 UP000242457 UP000005203 UP000053105 UP000053825 UP000092461 UP000078540 UP000005205 UP000078541 UP000000311 UP000007755 UP000279307 UP000030742 UP000078492 UP000053097 UP000078542 UP000002320 UP000215335 UP000075809 UP000075920 UP000030765 UP000075885 UP000008820 UP000076407 UP000183832 UP000075900 UP000075840 UP000075881 UP000069272 UP000075886 UP000076502 UP000075884 UP000002358 UP000019118 UP000007819 UP000009046 UP000069940 UP000249989 UP000076858 UP000000305 UP000075883 UP000054359 UP000076408
UP000007266 UP000235965 UP000242457 UP000005203 UP000053105 UP000053825 UP000092461 UP000078540 UP000005205 UP000078541 UP000000311 UP000007755 UP000279307 UP000030742 UP000078492 UP000053097 UP000078542 UP000002320 UP000215335 UP000075809 UP000075920 UP000030765 UP000075885 UP000008820 UP000076407 UP000183832 UP000075900 UP000075840 UP000075881 UP000069272 UP000075886 UP000076502 UP000075884 UP000002358 UP000019118 UP000007819 UP000009046 UP000069940 UP000249989 UP000076858 UP000000305 UP000075883 UP000054359 UP000076408
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IZG0
A0A2A4K869
A0A2W1BL44
A0A2H1VZM7
A0A212FJY1
A0A194PMV9
+ More
A0A194RH49 A0A0L7LV37 D6WRL2 A0A2J7RCF5 A0A2A3EIQ2 A0A0C9QT06 A0A088A5V6 A0A0M8ZTR7 A0A0L7R1T7 A0A1B0EVJ6 A0A195BAH8 A0A158NUV5 A0A151K0J4 E2A767 F4WE17 A0A3L8D6J8 U4UQG6 A0A151IV55 A0A026WD85 A0A151I6G1 B0W448 A0A232F4Y4 A0A151WP70 A0A182W8K3 A0A084WQ39 A0A182PM95 J9HSG8 A0A182XHA8 A0A1J1ISI4 A0A182R1R9 A0A182HR27 A0A182KH83 A0A182FN59 A0A182Q2D9 A0A154P3M5 A0A182NNU2 K7JNF7 A0A2S2NN49 N6U222 J9JKB5 E0VIC6 A0A182GXZ2 A0A2S2Q9Q2 A0A0N8CFH1 A0A0P5ND54 A0A0N8B5Q9 A0A0P5ZV18 A0A164NEH7 A0A0P5XRH6 A0A0P5XLF5 A0A0N8C3N1 A0A0N8B670 E9GBH0 A0A0P5D8W7 A0A0P6GPU8 A0A0P5ZV22 A0A0P6IMI2 A0A0P5SGW4 A0A0P5ZVI0 A0A0P5D981 A0A0P5K7C4 A0A0N8A4C8 A0A0N8DJ19 A0A0P6HRV6 A0A0P5LTZ7 A0A0P5RDW6 A0A0N8DHU4 A0A182M5L6 A0A0P4Z4M5 A0A0P5B1N8 A0A0N8AGF6 A0A0P6CYJ7 A0A087SYI7 A0A182Y641 A0A164NDJ1 A0A0K2TDM0
A0A194RH49 A0A0L7LV37 D6WRL2 A0A2J7RCF5 A0A2A3EIQ2 A0A0C9QT06 A0A088A5V6 A0A0M8ZTR7 A0A0L7R1T7 A0A1B0EVJ6 A0A195BAH8 A0A158NUV5 A0A151K0J4 E2A767 F4WE17 A0A3L8D6J8 U4UQG6 A0A151IV55 A0A026WD85 A0A151I6G1 B0W448 A0A232F4Y4 A0A151WP70 A0A182W8K3 A0A084WQ39 A0A182PM95 J9HSG8 A0A182XHA8 A0A1J1ISI4 A0A182R1R9 A0A182HR27 A0A182KH83 A0A182FN59 A0A182Q2D9 A0A154P3M5 A0A182NNU2 K7JNF7 A0A2S2NN49 N6U222 J9JKB5 E0VIC6 A0A182GXZ2 A0A2S2Q9Q2 A0A0N8CFH1 A0A0P5ND54 A0A0N8B5Q9 A0A0P5ZV18 A0A164NEH7 A0A0P5XRH6 A0A0P5XLF5 A0A0N8C3N1 A0A0N8B670 E9GBH0 A0A0P5D8W7 A0A0P6GPU8 A0A0P5ZV22 A0A0P6IMI2 A0A0P5SGW4 A0A0P5ZVI0 A0A0P5D981 A0A0P5K7C4 A0A0N8A4C8 A0A0N8DJ19 A0A0P6HRV6 A0A0P5LTZ7 A0A0P5RDW6 A0A0N8DHU4 A0A182M5L6 A0A0P4Z4M5 A0A0P5B1N8 A0A0N8AGF6 A0A0P6CYJ7 A0A087SYI7 A0A182Y641 A0A164NDJ1 A0A0K2TDM0
PDB
3B2Z
E-value=6.21248e-23,
Score=269
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
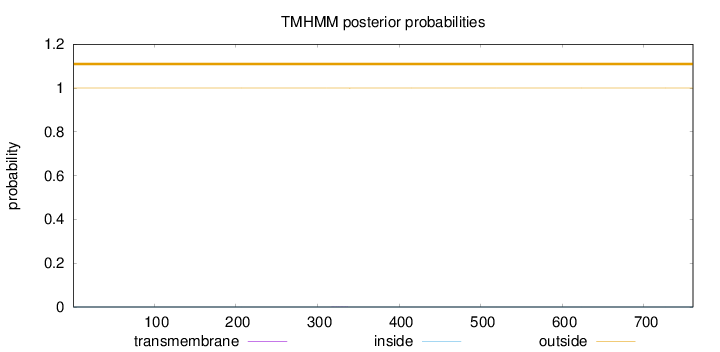
Length:
760
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00714
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00034
outside
1 - 760
Population Genetic Test Statistics
Pi
255.73108
Theta
165.426708
Tajima's D
1.269775
CLR
0.224186
CSRT
0.728063596820159
Interpretation
Uncertain
