Gene
KWMTBOMO02636
Pre Gene Modal
BGIBMGA002643
Annotation
PREDICTED:_phosphatidate_phosphatase_LPIN2_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.799
Sequence
CDS
ATGGGGTTGTACGGAAAACCGGCCGTCATGTATTCTATGAATTATATCGGTCAGTTCATAGCGAACTTTCGCGAATTCTATAATGAGATCAACGCAGCGACACTTACTGGAGCTATAGATGTCATCGTAGTCGAACAGCCCGATGGCAGCTTCACCTGTTCGCCATTCCACGTGCGTTTTGGGAAACTCGGCGTGCTTCGATCCAGATTTAAAGTGGTGGACCTAGAATTGAATGGTGAGCCTTTGAATATCCACATGAAGCTGGGGGAGTCTGGTGAGGCATTTTTCGTTGAGGAAGTAGGCGAAGACGAAGCTGAATGTTCCGCTCACCTTGCCACCTCGCCCATCCCGGCTGCAAGGTTCGAGGAATTATATGAGCCACGTCGTCGCAATTCGCTCTCTGCTGTCGAACCCGATCATGGGCAGGCATCAGATTATACTAAACGAAGATACACTGCAGACGGACAAACTATGCAGAAACCGGACTTGACCAAGATTAAGAAGAATACGACGAAAGACGAGAAACCATCAAAAGACAACGAGGCCCTGTTTGAAATGGAGGACCTCGACGAAGGTACCGACTGGACGGACGGAAAAACACCGAAAACGTTCGCGGCCACTCAGCTTGGCGAAGCGGAAAGCGAAGCGAATCAGGCCGTACAAAAAATTAGCGTCCACAATGACTTTAGACCTATAGACGTCGCGCCCACCGAGGAGGAGGTCAAGCAGAATGGGAACGGCAAGAAAAAGAAAAAAACCAGAAAAGTCAGCTCGAAGAAGAAGCACCAAAGAAAGTCGTCCACTAGTTCTGTATCTAGTCAGTCGGACCGCGCTGGCTCGGAGCAACGAGCCTTGCAGTCGACCGCCATTGTTAACGCCACTGATATGAGTTTCTTTAGTGACACCGAACTGAATGCTGCTACATTGAGCAGAGGTTCTCGGGGCGGTACTCCAGTACAGTCGGACACCGAGGTGGAGCTTTCGAGAACAACGACCGGCGACAAGTCATCGCAGTCGTGGAGGTGGGGCGAACTCCCGGAGCCTCCGGTGCGCGGGGTGGAGTCAGGCGAGTGCGCGTCGGCGCCGTCCCCTGCGTCCGCCTCCCCCGCGTCCCCCTCCTCGCCCGCTTCCCCCGCCTCGCCCGCCGAGCCGCTCAGCTCCGACGAGGAGCGGCCCAGCCGGCGTGGCGTGCTCAGCGGCATGTTCCGCTTCATGTCGACGCGGGACTCTGCCGACAACGCGCCCGAGGGGATCTACCTGGACGACCTGGACCGTGGTCAGGTCAACCCCGACCTGTACTTCCCGAAATATCATGCTGAACGTCATCGTGGCAGCTCGGTGGTGCAGCACGCGCGCGCCGCTCCCACGGAGGAGGAGTACGAGTCCGGGAACGGCCCGTCCCTGCCGCAGTCGCCAGCCTCGCTCGAACCGCCCGTCATGGACTCGGACGACGACGACAAAATCTTAGCAAAGGGTCAAGTGTGTCTGTACGTGCGCGAGGACAACGTGTCCCGCGCGCTGCCCTTCGAGGAGTTCTGCTCCGACGGCGCCCGACTGGCGGCGGAGGGCCGGCTGCTGGTGCGGCTCGGCGACCGCTGGATGCCGTGGCGCAGCGCGGGGCCGCTCATCCTGTCCCTGCTCGTGTACCGCCGGCCTTTGCCAAACCGCGTCTTGGAGTCGCTAGAGAAAGGTTCGGAGAAGGAAGAGGCGGACGCGGCGAGCTCGCACCGCGGAGACAGCGGCAGCAAGCCCCGCGCCTACTCCTGGTGGAGCTGGAGGCGGACGTCCGAGGCTAAACATGCCGAAAAGACTTCTCTATCGGACGACATTCCAACAAAGTCGAGTCCACCACAATCCGAAGCCGTAGACACTGTTGCACTCCAAGAGACGCCTGCGGAAGAATCGACTGTGGAAACAATCGAAGAGACACAAAACATTGTAAACGTTGATGACGTGTTCGAATCTCAGCCCGTCGAAGTGGAGGAAAACATACTTACGATGTCAGAAAACGCTGTCCACCAAGAACAAGCAATGGATCACCAAGACCAGAGTTCATCGGATTCGGATCAAGAGCAGCAGCTGCCCAGGTCGTCGAGGGCGCGCCGCCACTCGACCTTCCGGAAGACCCTTCGTTTGTCATCAGAACAGATTAGAAACTTGAACCTCCGCGAGGGCATGAACGAGATGGTGTTCAGCGTGACCACGGCGTACCAGGGCACCACGCGCTGCAAGTGCAACGTGTTCCGCTGGCGATACGACGACAAGGTCGTCATCTCCGACATCGACGGAACCATCACCAAATCGGACGTACTGGGCCATATATTCCCAATGGTTGGCAAAGATTGGGCCCAGTCTGGGGTCGCTCAACTATTCACGAAGATAAAGAACAACGGATATCAACTGCTCTATCTGTCGGCCAGGGCTATCGGACAAGCCAAAGTGACTCGGGAATATCTGCGCTCAATCAGACAAGGCGAGGTGTGTCTACCGGACGGGCCGCTGCTGCTGAACCCTACGTCGTTGCTTCGCGCCTTCCATCGAGAGGTCATCGAGAAGAAACCAGAAGAATTCAAAATTCAGTGCCTCGCCGATATCAAGGCGCTGTTCCCTCAAGGGTCTAATCCATTTTACGCTGGCTATGGGAATAGAGTGAATGACGTTTGTGCATATCAAGCTGTCGGTATTCCGATTGTAAGAATTTTCACCATAAACTATAAGGGCGAATTAAAACATGAACTAACACAGACATTCCAGTCCACGTATTCCCATATGTCAGTGCTGGTGGACCAGGTGTTTCCGCCGGCGCAGTGCGAGCCCAGCGACGAGTTCTCGCAAACGGTGTACTGGCGCGACCCGCTGCCCGCCGTCGACCTGCCACCGATCGTCCCACCGCAGCCCAAACATTAA
Protein
MGLYGKPAVMYSMNYIGQFIANFREFYNEINAATLTGAIDVIVVEQPDGSFTCSPFHVRFGKLGVLRSRFKVVDLELNGEPLNIHMKLGESGEAFFVEEVGEDEAECSAHLATSPIPAARFEELYEPRRRNSLSAVEPDHGQASDYTKRRYTADGQTMQKPDLTKIKKNTTKDEKPSKDNEALFEMEDLDEGTDWTDGKTPKTFAATQLGEAESEANQAVQKISVHNDFRPIDVAPTEEEVKQNGNGKKKKKTRKVSSKKKHQRKSSTSSVSSQSDRAGSEQRALQSTAIVNATDMSFFSDTELNAATLSRGSRGGTPVQSDTEVELSRTTTGDKSSQSWRWGELPEPPVRGVESGECASAPSPASASPASPSSPASPASPAEPLSSDEERPSRRGVLSGMFRFMSTRDSADNAPEGIYLDDLDRGQVNPDLYFPKYHAERHRGSSVVQHARAAPTEEEYESGNGPSLPQSPASLEPPVMDSDDDDKILAKGQVCLYVREDNVSRALPFEEFCSDGARLAAEGRLLVRLGDRWMPWRSAGPLILSLLVYRRPLPNRVLESLEKGSEKEEADAASSHRGDSGSKPRAYSWWSWRRTSEAKHAEKTSLSDDIPTKSSPPQSEAVDTVALQETPAEESTVETIEETQNIVNVDDVFESQPVEVEENILTMSENAVHQEQAMDHQDQSSSDSDQEQQLPRSSRARRHSTFRKTLRLSSEQIRNLNLREGMNEMVFSVTTAYQGTTRCKCNVFRWRYDDKVVISDIDGTITKSDVLGHIFPMVGKDWAQSGVAQLFTKIKNNGYQLLYLSARAIGQAKVTREYLRSIRQGEVCLPDGPLLLNPTSLLRAFHREVIEKKPEEFKIQCLADIKALFPQGSNPFYAGYGNRVNDVCAYQAVGIPIVRIFTINYKGELKHELTQTFQSTYSHMSVLVDQVFPPAQCEPSDEFSQTVYWRDPLPAVDLPPIVPPQPKH
Summary
Uniprot
H9IZF8
A0A2A4K7D8
A0A194PUE7
A0A2A4K6Z7
A0A2A4K6I6
A0A212FJV5
+ More
A0A2A4K7V5 A0A2H1VZL6 A0A1E1W7Z0 A0A2W1BLJ4 D6WRL3 U4UQ71 N6TZG0 E2BMP7 A0A151JSX5 A0A088AHG4 A0A310SD58 A0A2A3EA97 V9IC47 A0A158NNY6 A0A195EX40 A0A0L7RJ49 A0A195BRZ0 A0A151WJR2 A0A151I741 A0A0M8ZR02 A0A067RVD1 A0A336MZ72 A0A336MY31 A0A154NZL2 E2A3I3 A0A0C9QZY0 V9IBP7 A0A026WQV2 A0A1B6KH09 V5GVQ9 A0A3L8DNH2 A0A1B6KA32 A0A034V710 W8BJ41 A0A1I8P291 A0A0K8WEI7 A0A034V5X6 A0A0Q9W5A0 A0A0Q9XHU0 A0A069DX92 A0A1B6KFH0 A0A162PAC4 A0A1I8MAI8 A0A0P5FGT4 A0A0P5LIW4 A0A0P4Y8N3 A0A0P5RL61 A0A0P5L4A7 A0A023F323 A0A0P5UT05 A0A0P5K5W5 A0A0P5S311 A0A0P4VVG7 A0A0P5M5Z3 A0A0P5QL63 A0A0P5V1A9 A0A1B6DCG0 A0A1B6EBL2 A0A0P5JXC0 A0A0M4ELJ9 A0A0P5Q222 A0A0N7ZLN4 A0A224X6N6 A0A0P5ZKD5 A0A0P5WL16 A0A0P4YBY6 A0A0P5UXG7 A0A1B6LEL7 A0A0P4ZAN7 A0A0P5Y338 A0A0P5W1W3 A0A0P5V6F3 A0A0P5IXL4 A0A0P5FX13 A0A1A9XB69 A0A1B0BV46 A0A0P5IRU2 A0A0N8BSF0
A0A2A4K7V5 A0A2H1VZL6 A0A1E1W7Z0 A0A2W1BLJ4 D6WRL3 U4UQ71 N6TZG0 E2BMP7 A0A151JSX5 A0A088AHG4 A0A310SD58 A0A2A3EA97 V9IC47 A0A158NNY6 A0A195EX40 A0A0L7RJ49 A0A195BRZ0 A0A151WJR2 A0A151I741 A0A0M8ZR02 A0A067RVD1 A0A336MZ72 A0A336MY31 A0A154NZL2 E2A3I3 A0A0C9QZY0 V9IBP7 A0A026WQV2 A0A1B6KH09 V5GVQ9 A0A3L8DNH2 A0A1B6KA32 A0A034V710 W8BJ41 A0A1I8P291 A0A0K8WEI7 A0A034V5X6 A0A0Q9W5A0 A0A0Q9XHU0 A0A069DX92 A0A1B6KFH0 A0A162PAC4 A0A1I8MAI8 A0A0P5FGT4 A0A0P5LIW4 A0A0P4Y8N3 A0A0P5RL61 A0A0P5L4A7 A0A023F323 A0A0P5UT05 A0A0P5K5W5 A0A0P5S311 A0A0P4VVG7 A0A0P5M5Z3 A0A0P5QL63 A0A0P5V1A9 A0A1B6DCG0 A0A1B6EBL2 A0A0P5JXC0 A0A0M4ELJ9 A0A0P5Q222 A0A0N7ZLN4 A0A224X6N6 A0A0P5ZKD5 A0A0P5WL16 A0A0P4YBY6 A0A0P5UXG7 A0A1B6LEL7 A0A0P4ZAN7 A0A0P5Y338 A0A0P5W1W3 A0A0P5V6F3 A0A0P5IXL4 A0A0P5FX13 A0A1A9XB69 A0A1B0BV46 A0A0P5IRU2 A0A0N8BSF0
Pubmed
EMBL
BABH01014136
BABH01014137
NWSH01000078
PCG79848.1
KQ459597
KPI94760.1
+ More
PCG79849.1 PCG79851.1 AGBW02008206 OWR54023.1 PCG79853.1 ODYU01005438 SOQ46285.1 GDQN01007985 JAT83069.1 KZ150098 PZC73580.1 KQ971354 EFA07054.2 KB632319 ERL92286.1 APGK01045628 KB741037 ENN74670.1 GL449285 EFN83032.1 KQ978538 KYN30251.1 KQ775960 OAD52223.1 KZ288310 PBC28638.1 JR037942 AEY58046.1 ADTU01021786 KQ981953 KYN32467.1 KQ414583 KOC70776.1 KQ976417 KYM89797.1 KQ983031 KYQ48109.1 KQ978438 KYM93964.1 KQ435969 KOX67863.1 KK852442 KDR23839.1 UFQS01002555 UFQT01002555 SSX14262.1 SSX33677.1 UFQT01003532 SSX35090.1 KQ434786 KZC05109.1 GL436428 EFN72047.1 GBYB01006247 GBYB01006485 JAG76014.1 JAG76252.1 JR037943 AEY58047.1 KK107128 EZA58410.1 GEBQ01029251 JAT10726.1 GALX01004138 JAB64328.1 QOIP01000006 RLU21957.1 GEBQ01031669 JAT08308.1 GAKP01021412 JAC37540.1 GAMC01016947 GAMC01016946 GAMC01016945 GAMC01016943 JAB89609.1 GDHF01002726 JAI49588.1 GAKP01021411 GAKP01021410 JAC37542.1 CH940648 KRF80093.1 CH933808 KRG04414.1 GBGD01000374 JAC88515.1 GEBQ01029808 JAT10169.1 LRGB01000512 KZS18674.1 GDIP01148083 JAJ75319.1 GDIQ01168960 JAK82765.1 GDIP01233262 JAI90139.1 GDIQ01099176 JAL52550.1 GDIQ01200144 JAK51581.1 GBBI01003201 JAC15511.1 GDIP01108777 JAL94937.1 GDIQ01190301 JAK61424.1 GDIQ01102079 JAL49647.1 GDKW01000029 JAI56566.1 GDIQ01184254 JAK67471.1 GDIQ01115672 JAL36054.1 GDIP01106376 JAL97338.1 GEDC01031379 GEDC01017183 GEDC01013925 GEDC01011810 JAS05919.1 JAS20115.1 JAS23373.1 JAS25488.1 GEDC01002034 GEDC01000644 JAS35264.1 JAS36654.1 GDIQ01192192 JAK59533.1 CP012524 ALC42467.1 GDIQ01120221 JAL31505.1 GDIP01233265 JAI90136.1 GFTR01008301 JAW08125.1 GDIP01055936 JAM47779.1 GDIP01084780 JAM18935.1 GDIP01229677 JAI93724.1 GDIP01108779 JAL94935.1 GEBQ01017845 JAT22132.1 GDIP01215514 JAJ07888.1 GDIP01063693 JAM40022.1 GDIP01106377 JAL97337.1 GDIP01106378 JAL97336.1 GDIQ01206190 JAK45535.1 GDIQ01250477 JAK01248.1 JXJN01021142 GDIQ01208850 JAK42875.1 GDIQ01149459 JAL02267.1
PCG79849.1 PCG79851.1 AGBW02008206 OWR54023.1 PCG79853.1 ODYU01005438 SOQ46285.1 GDQN01007985 JAT83069.1 KZ150098 PZC73580.1 KQ971354 EFA07054.2 KB632319 ERL92286.1 APGK01045628 KB741037 ENN74670.1 GL449285 EFN83032.1 KQ978538 KYN30251.1 KQ775960 OAD52223.1 KZ288310 PBC28638.1 JR037942 AEY58046.1 ADTU01021786 KQ981953 KYN32467.1 KQ414583 KOC70776.1 KQ976417 KYM89797.1 KQ983031 KYQ48109.1 KQ978438 KYM93964.1 KQ435969 KOX67863.1 KK852442 KDR23839.1 UFQS01002555 UFQT01002555 SSX14262.1 SSX33677.1 UFQT01003532 SSX35090.1 KQ434786 KZC05109.1 GL436428 EFN72047.1 GBYB01006247 GBYB01006485 JAG76014.1 JAG76252.1 JR037943 AEY58047.1 KK107128 EZA58410.1 GEBQ01029251 JAT10726.1 GALX01004138 JAB64328.1 QOIP01000006 RLU21957.1 GEBQ01031669 JAT08308.1 GAKP01021412 JAC37540.1 GAMC01016947 GAMC01016946 GAMC01016945 GAMC01016943 JAB89609.1 GDHF01002726 JAI49588.1 GAKP01021411 GAKP01021410 JAC37542.1 CH940648 KRF80093.1 CH933808 KRG04414.1 GBGD01000374 JAC88515.1 GEBQ01029808 JAT10169.1 LRGB01000512 KZS18674.1 GDIP01148083 JAJ75319.1 GDIQ01168960 JAK82765.1 GDIP01233262 JAI90139.1 GDIQ01099176 JAL52550.1 GDIQ01200144 JAK51581.1 GBBI01003201 JAC15511.1 GDIP01108777 JAL94937.1 GDIQ01190301 JAK61424.1 GDIQ01102079 JAL49647.1 GDKW01000029 JAI56566.1 GDIQ01184254 JAK67471.1 GDIQ01115672 JAL36054.1 GDIP01106376 JAL97338.1 GEDC01031379 GEDC01017183 GEDC01013925 GEDC01011810 JAS05919.1 JAS20115.1 JAS23373.1 JAS25488.1 GEDC01002034 GEDC01000644 JAS35264.1 JAS36654.1 GDIQ01192192 JAK59533.1 CP012524 ALC42467.1 GDIQ01120221 JAL31505.1 GDIP01233265 JAI90136.1 GFTR01008301 JAW08125.1 GDIP01055936 JAM47779.1 GDIP01084780 JAM18935.1 GDIP01229677 JAI93724.1 GDIP01108779 JAL94935.1 GEBQ01017845 JAT22132.1 GDIP01215514 JAJ07888.1 GDIP01063693 JAM40022.1 GDIP01106377 JAL97337.1 GDIP01106378 JAL97336.1 GDIQ01206190 JAK45535.1 GDIQ01250477 JAK01248.1 JXJN01021142 GDIQ01208850 JAK42875.1 GDIQ01149459 JAL02267.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000007266
UP000030742
+ More
UP000019118 UP000008237 UP000078492 UP000005203 UP000242457 UP000005205 UP000078541 UP000053825 UP000078540 UP000075809 UP000078542 UP000053105 UP000027135 UP000076502 UP000000311 UP000053097 UP000279307 UP000095300 UP000008792 UP000009192 UP000076858 UP000095301 UP000092553 UP000092443 UP000092460
UP000019118 UP000008237 UP000078492 UP000005203 UP000242457 UP000005205 UP000078541 UP000053825 UP000078540 UP000075809 UP000078542 UP000053105 UP000027135 UP000076502 UP000000311 UP000053097 UP000279307 UP000095300 UP000008792 UP000009192 UP000076858 UP000095301 UP000092553 UP000092443 UP000092460
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IZF8
A0A2A4K7D8
A0A194PUE7
A0A2A4K6Z7
A0A2A4K6I6
A0A212FJV5
+ More
A0A2A4K7V5 A0A2H1VZL6 A0A1E1W7Z0 A0A2W1BLJ4 D6WRL3 U4UQ71 N6TZG0 E2BMP7 A0A151JSX5 A0A088AHG4 A0A310SD58 A0A2A3EA97 V9IC47 A0A158NNY6 A0A195EX40 A0A0L7RJ49 A0A195BRZ0 A0A151WJR2 A0A151I741 A0A0M8ZR02 A0A067RVD1 A0A336MZ72 A0A336MY31 A0A154NZL2 E2A3I3 A0A0C9QZY0 V9IBP7 A0A026WQV2 A0A1B6KH09 V5GVQ9 A0A3L8DNH2 A0A1B6KA32 A0A034V710 W8BJ41 A0A1I8P291 A0A0K8WEI7 A0A034V5X6 A0A0Q9W5A0 A0A0Q9XHU0 A0A069DX92 A0A1B6KFH0 A0A162PAC4 A0A1I8MAI8 A0A0P5FGT4 A0A0P5LIW4 A0A0P4Y8N3 A0A0P5RL61 A0A0P5L4A7 A0A023F323 A0A0P5UT05 A0A0P5K5W5 A0A0P5S311 A0A0P4VVG7 A0A0P5M5Z3 A0A0P5QL63 A0A0P5V1A9 A0A1B6DCG0 A0A1B6EBL2 A0A0P5JXC0 A0A0M4ELJ9 A0A0P5Q222 A0A0N7ZLN4 A0A224X6N6 A0A0P5ZKD5 A0A0P5WL16 A0A0P4YBY6 A0A0P5UXG7 A0A1B6LEL7 A0A0P4ZAN7 A0A0P5Y338 A0A0P5W1W3 A0A0P5V6F3 A0A0P5IXL4 A0A0P5FX13 A0A1A9XB69 A0A1B0BV46 A0A0P5IRU2 A0A0N8BSF0
A0A2A4K7V5 A0A2H1VZL6 A0A1E1W7Z0 A0A2W1BLJ4 D6WRL3 U4UQ71 N6TZG0 E2BMP7 A0A151JSX5 A0A088AHG4 A0A310SD58 A0A2A3EA97 V9IC47 A0A158NNY6 A0A195EX40 A0A0L7RJ49 A0A195BRZ0 A0A151WJR2 A0A151I741 A0A0M8ZR02 A0A067RVD1 A0A336MZ72 A0A336MY31 A0A154NZL2 E2A3I3 A0A0C9QZY0 V9IBP7 A0A026WQV2 A0A1B6KH09 V5GVQ9 A0A3L8DNH2 A0A1B6KA32 A0A034V710 W8BJ41 A0A1I8P291 A0A0K8WEI7 A0A034V5X6 A0A0Q9W5A0 A0A0Q9XHU0 A0A069DX92 A0A1B6KFH0 A0A162PAC4 A0A1I8MAI8 A0A0P5FGT4 A0A0P5LIW4 A0A0P4Y8N3 A0A0P5RL61 A0A0P5L4A7 A0A023F323 A0A0P5UT05 A0A0P5K5W5 A0A0P5S311 A0A0P4VVG7 A0A0P5M5Z3 A0A0P5QL63 A0A0P5V1A9 A0A1B6DCG0 A0A1B6EBL2 A0A0P5JXC0 A0A0M4ELJ9 A0A0P5Q222 A0A0N7ZLN4 A0A224X6N6 A0A0P5ZKD5 A0A0P5WL16 A0A0P4YBY6 A0A0P5UXG7 A0A1B6LEL7 A0A0P4ZAN7 A0A0P5Y338 A0A0P5W1W3 A0A0P5V6F3 A0A0P5IXL4 A0A0P5FX13 A0A1A9XB69 A0A1B0BV46 A0A0P5IRU2 A0A0N8BSF0
Ontologies
PATHWAY
PANTHER
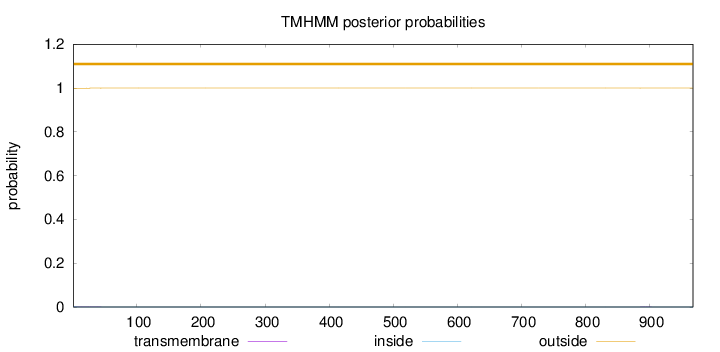
Topology
Length:
968
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02468
Exp number, first 60 AAs:
0.01899
Total prob of N-in:
0.00097
outside
1 - 968
Population Genetic Test Statistics
Pi
235.223511
Theta
184.388082
Tajima's D
0.47162
CLR
1.005342
CSRT
0.506874656267187
Interpretation
Uncertain
