Gene
KWMTBOMO02635
Pre Gene Modal
BGIBMGA002642
Annotation
PREDICTED:_ras-related_GTP-binding_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.287
Sequence
CDS
ATGGACGATCCAAATTGTTATGTGGGGTCGTTCCCCAAAGATTTTACCTATGGACCTTTCGAGCAAAATGGAGATGGCGATAGTTCTTCTCTTCAAGACGATCACAAACCACGAATCCTTTTAATGGGCCTTAGAAGGTCTGGAAAATCATCAATTCAAAAAGTTGTATTTCACAAGATGTCTCCAAATGAGACATTGTTTTTAGAATCTACAAATCAAATTGTTAAAGACGACATTAATAACAGCAGTTTTGTACAATTTCAAATATGGGACTTTCCCGGGCAGATAGACTTCTTTGATCCTACATTTGATTCTGACACCATATTTGGTGGATGTGGAGCTCTTGTCTTTGTAATTGATGCGCAGGATGATTATCAAGATGCTTTGGATAAATTACAACTTACTGTCACTAAAGCCTACAGAGTGAATAGTAATATTAAATTTGAGGTGTTCATACATAAAGTTGATGGTCTCAATGATGATTACAAAATGGAATCTCAAAGGGATATACACCACCGGGCCACTGATGATCTTGCAGAAGCTGGTCTCCATCATGTACATCTATCCTTCCATCTTACAAGCATATATGACCATTCAATATTTGAAGCATTTAGCAAAGTAGTCCAAAAGCTAATACCTCAGTTGCCTACATTAGAAAATCTTTTAAATATACTGATATCTAATTCCGGAATAGAAAAGGCATTCCTATTTGATGTAGTATCAAAGATTTATATAGCCACTGACAGTTCACCAGTAGATATGCAAAGCTATGAGCTTTGCTGTGATATGATTGATGTCGTAATTGATATATCTTGTATTTATGGCATGGTGGATGAAACTGAAACAAATACGTTCAACAATGAAAGTTCGAGTTTGATAAAACTAAACAATGGTACTGTACTATATCTACGAGAGGTCAACAAATTCTTGGCATTGGTTTGCATTCTCAGGGAAGAAAATTTCCAAAAACAAGGTGTGATCGATTACAATTTCTTATGTTTCCGGCATGCTATTACGCAAGTCTTTGAACTTCGTAATAAATGTCAAAGTTCTGGAACTGACAGAAGAAGATCGGGTACTCCAGATGCTGTTCCAAACGGTGCTGCAGATTCCACTGAGAGTACTTCAGATCATTCACAGCCTCAGTTAGCATAA
Protein
MDDPNCYVGSFPKDFTYGPFEQNGDGDSSSLQDDHKPRILLMGLRRSGKSSIQKVVFHKMSPNETLFLESTNQIVKDDINNSSFVQFQIWDFPGQIDFFDPTFDSDTIFGGCGALVFVIDAQDDYQDALDKLQLTVTKAYRVNSNIKFEVFIHKVDGLNDDYKMESQRDIHHRATDDLAEAGLHHVHLSFHLTSIYDHSIFEAFSKVVQKLIPQLPTLENLLNILISNSGIEKAFLFDVVSKIYIATDSSPVDMQSYELCCDMIDVVIDISCIYGMVDETETNTFNNESSSLIKLNNGTVLYLREVNKFLALVCILREENFQKQGVIDYNFLCFRHAITQVFELRNKCQSSGTDRRRSGTPDAVPNGAADSTESTSDHSQPQLA
Summary
Uniprot
D3JSV8
A0A2W1BEM8
A0A2A4K859
A0A1E1WRI6
A0A194RFP6
A0A194PPU8
+ More
A0A212FJW8 S4PYB6 H9IZF7 K7J4B1 A0A067RSQ5 A0A2J7RG12 A0A2J7RG05 A0A2J7RG15 E2BKQ2 A0A232F829 E2APE4 A0A2A3E2G3 A0A088ASD0 A0A026VVY8 E9I9Q3 A0A158NTD3 A0A195CLT5 A0A151J9X9 F4W3Y3 B0W3P3 A0A023ERA1 A0A1Q3FEE5 A0A1Y1KMZ7 A0A0N1ITX8 A0A1Y1KMC1 Q17AT2 A0A1B6CL71 A0A1B6C546 A0A1B6H0Y2 A0A1B6INS3 A0A1L8DVF6 A0A182QCF8 A0A1B6M7G7 T1JL47 A0A1S3I7K2 A0A182KRU5 A0A182UDT4 A0A182UVR9 Q7PTD6 A0A182I3Q2 A0A182X6E7 U5EQE9 A0A182JNY2 A0A182MQM4 A0A182NHK8 A0A182XZ98 A0A151X1S2 A0A195B3U0 A0A0T6BAL1 A0A154NZZ5 A0A182RFP1 A0A182WIS2 A0A2B4R6R1 A0A182P3J1 A0A1B0GJ67 W5J9P0 A0A2M4A7C1 A0A0M4MP51 A0A1B0D5F4 A0A2M4BR90 A0A2M3Z4X6 A0A034VEC9 W8BV62 A0A0K8VIP9 A0A1I8PBQ1 A0A1W4WL25 A0A182JFE4 W4ZAA5 A0A0K8TPU2 A0A0L0BPL7 D3TPQ6 T1PC66 B4J8C5 A7SI02 Q28ZX7 D6WDF9 E0VN58 A0A3B0JPG1 N6TKD7 A0A0L8FRD0 A0A2G8LCZ1 A0A1W4WB83 B4P2P3 B3MGU0 A0A1A9X4A0 A0A0J9R963 A0A0L7R403 Q7K519 B4QFB9 B4HRK9 B4LJN3 A0A2T7PF46 A0A087U2C6 B3N8T4
A0A212FJW8 S4PYB6 H9IZF7 K7J4B1 A0A067RSQ5 A0A2J7RG12 A0A2J7RG05 A0A2J7RG15 E2BKQ2 A0A232F829 E2APE4 A0A2A3E2G3 A0A088ASD0 A0A026VVY8 E9I9Q3 A0A158NTD3 A0A195CLT5 A0A151J9X9 F4W3Y3 B0W3P3 A0A023ERA1 A0A1Q3FEE5 A0A1Y1KMZ7 A0A0N1ITX8 A0A1Y1KMC1 Q17AT2 A0A1B6CL71 A0A1B6C546 A0A1B6H0Y2 A0A1B6INS3 A0A1L8DVF6 A0A182QCF8 A0A1B6M7G7 T1JL47 A0A1S3I7K2 A0A182KRU5 A0A182UDT4 A0A182UVR9 Q7PTD6 A0A182I3Q2 A0A182X6E7 U5EQE9 A0A182JNY2 A0A182MQM4 A0A182NHK8 A0A182XZ98 A0A151X1S2 A0A195B3U0 A0A0T6BAL1 A0A154NZZ5 A0A182RFP1 A0A182WIS2 A0A2B4R6R1 A0A182P3J1 A0A1B0GJ67 W5J9P0 A0A2M4A7C1 A0A0M4MP51 A0A1B0D5F4 A0A2M4BR90 A0A2M3Z4X6 A0A034VEC9 W8BV62 A0A0K8VIP9 A0A1I8PBQ1 A0A1W4WL25 A0A182JFE4 W4ZAA5 A0A0K8TPU2 A0A0L0BPL7 D3TPQ6 T1PC66 B4J8C5 A7SI02 Q28ZX7 D6WDF9 E0VN58 A0A3B0JPG1 N6TKD7 A0A0L8FRD0 A0A2G8LCZ1 A0A1W4WB83 B4P2P3 B3MGU0 A0A1A9X4A0 A0A0J9R963 A0A0L7R403 Q7K519 B4QFB9 B4HRK9 B4LJN3 A0A2T7PF46 A0A087U2C6 B3N8T4
Pubmed
28756777
26354079
22118469
23622113
19121390
20075255
+ More
24845553 20798317 28648823 24508170 30249741 21282665 21347285 21719571 24945155 28004739 17510324 20966253 12364791 14747013 17210077 25244985 20920257 23761445 25348373 24495485 26369729 26108605 20353571 25315136 17994087 17615350 15632085 18362917 19820115 20566863 23537049 29023486 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
24845553 20798317 28648823 24508170 30249741 21282665 21347285 21719571 24945155 28004739 17510324 20966253 12364791 14747013 17210077 25244985 20920257 23761445 25348373 24495485 26369729 26108605 20353571 25315136 17994087 17615350 15632085 18362917 19820115 20566863 23537049 29023486 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
GU288815
ADB78702.1
KZ150098
PZC73582.1
NWSH01000078
PCG79842.1
+ More
GDQN01001415 JAT89639.1 KQ460207 KPJ16643.1 KQ459597 KPI94759.1 AGBW02008206 OWR54024.1 GAIX01003573 JAA88987.1 BABH01014139 BABH01014140 AAZX01009455 KK852442 KDR23840.1 NEVH01004407 PNF39762.1 PNF39761.1 PNF39760.1 GL448819 EFN83750.1 NNAY01000718 OXU26815.1 GL441524 EFN64689.1 KZ288425 PBC25933.1 KK107796 QOIP01000013 EZA47651.1 RLU15580.1 GL761846 EFZ22703.1 ADTU01025845 KQ977642 KYN01034.1 KQ979352 KYN21844.1 GL887491 EGI70979.1 DS231833 EDS32190.1 GAPW01001898 JAC11700.1 GFDL01009121 JAV25924.1 GEZM01084049 JAV60816.1 KQ435726 KOX78128.1 GEZM01084050 JAV60815.1 CH477330 EAT43376.1 GEDC01023122 JAS14176.1 GEDC01028672 JAS08626.1 GECZ01001444 JAS68325.1 GECU01019131 JAS88575.1 GFDF01003660 JAV10424.1 AXCN02000593 GEBQ01008090 JAT31887.1 JH431827 AAAB01008807 EAA04060.5 APCN01000217 GANO01003294 JAB56577.1 AXCM01000671 KQ982585 KYQ54216.1 KQ976625 KYM78937.1 LJIG01002570 KRT84370.1 KQ434786 KZC05181.1 LSMT01001334 PFX12499.1 AJWK01019416 ADMH02001751 ETN61182.1 GGFK01003363 MBW36684.1 KR075857 ALE20576.1 AJVK01025173 GGFJ01006455 MBW55596.1 GGFM01002830 MBW23581.1 GAKP01018490 GAKP01018489 JAC40462.1 GAMC01013561 GAMC01013559 JAB92994.1 GDHF01013581 JAI38733.1 AAGJ04111141 AAGJ04111142 AAGJ04111143 GDAI01001688 JAI15915.1 JRES01001567 KNC21908.1 CCAG010004272 EZ423408 ADD19684.1 KA646356 AFP60985.1 CH916367 EDW02284.1 DS469664 EDO36641.1 CM000071 EAL25485.3 KQ971322 EEZ99978.2 DS235332 EEB14778.1 OUUW01000001 SPP75226.1 APGK01034587 KB740914 KB632352 ENN78378.1 ERL93255.1 KQ427255 KOF67271.1 MRZV01000121 PIK58129.1 CM000157 EDW89304.2 CH902619 EDV37858.1 CM002911 KMY92219.1 KQ414663 KOC65481.1 AE013599 AY051510 AAF59120.2 AAK92934.1 CM000362 EDX06148.1 CH480816 EDW46891.1 CH940648 EDW61601.1 PZQS01000004 PVD32042.1 KK117832 KFM71515.1 CH954177 EDV59561.1
GDQN01001415 JAT89639.1 KQ460207 KPJ16643.1 KQ459597 KPI94759.1 AGBW02008206 OWR54024.1 GAIX01003573 JAA88987.1 BABH01014139 BABH01014140 AAZX01009455 KK852442 KDR23840.1 NEVH01004407 PNF39762.1 PNF39761.1 PNF39760.1 GL448819 EFN83750.1 NNAY01000718 OXU26815.1 GL441524 EFN64689.1 KZ288425 PBC25933.1 KK107796 QOIP01000013 EZA47651.1 RLU15580.1 GL761846 EFZ22703.1 ADTU01025845 KQ977642 KYN01034.1 KQ979352 KYN21844.1 GL887491 EGI70979.1 DS231833 EDS32190.1 GAPW01001898 JAC11700.1 GFDL01009121 JAV25924.1 GEZM01084049 JAV60816.1 KQ435726 KOX78128.1 GEZM01084050 JAV60815.1 CH477330 EAT43376.1 GEDC01023122 JAS14176.1 GEDC01028672 JAS08626.1 GECZ01001444 JAS68325.1 GECU01019131 JAS88575.1 GFDF01003660 JAV10424.1 AXCN02000593 GEBQ01008090 JAT31887.1 JH431827 AAAB01008807 EAA04060.5 APCN01000217 GANO01003294 JAB56577.1 AXCM01000671 KQ982585 KYQ54216.1 KQ976625 KYM78937.1 LJIG01002570 KRT84370.1 KQ434786 KZC05181.1 LSMT01001334 PFX12499.1 AJWK01019416 ADMH02001751 ETN61182.1 GGFK01003363 MBW36684.1 KR075857 ALE20576.1 AJVK01025173 GGFJ01006455 MBW55596.1 GGFM01002830 MBW23581.1 GAKP01018490 GAKP01018489 JAC40462.1 GAMC01013561 GAMC01013559 JAB92994.1 GDHF01013581 JAI38733.1 AAGJ04111141 AAGJ04111142 AAGJ04111143 GDAI01001688 JAI15915.1 JRES01001567 KNC21908.1 CCAG010004272 EZ423408 ADD19684.1 KA646356 AFP60985.1 CH916367 EDW02284.1 DS469664 EDO36641.1 CM000071 EAL25485.3 KQ971322 EEZ99978.2 DS235332 EEB14778.1 OUUW01000001 SPP75226.1 APGK01034587 KB740914 KB632352 ENN78378.1 ERL93255.1 KQ427255 KOF67271.1 MRZV01000121 PIK58129.1 CM000157 EDW89304.2 CH902619 EDV37858.1 CM002911 KMY92219.1 KQ414663 KOC65481.1 AE013599 AY051510 AAF59120.2 AAK92934.1 CM000362 EDX06148.1 CH480816 EDW46891.1 CH940648 EDW61601.1 PZQS01000004 PVD32042.1 KK117832 KFM71515.1 CH954177 EDV59561.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000002358
+ More
UP000027135 UP000235965 UP000008237 UP000215335 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000005205 UP000078542 UP000078492 UP000007755 UP000002320 UP000053105 UP000008820 UP000075886 UP000085678 UP000075882 UP000075902 UP000075903 UP000007062 UP000075840 UP000076407 UP000075881 UP000075883 UP000075884 UP000076408 UP000075809 UP000078540 UP000076502 UP000075900 UP000075920 UP000225706 UP000075885 UP000092461 UP000000673 UP000092462 UP000095300 UP000192223 UP000075880 UP000007110 UP000037069 UP000092444 UP000095301 UP000001070 UP000001593 UP000001819 UP000007266 UP000009046 UP000268350 UP000019118 UP000030742 UP000053454 UP000230750 UP000192221 UP000002282 UP000007801 UP000091820 UP000053825 UP000000803 UP000000304 UP000001292 UP000008792 UP000245119 UP000054359 UP000008711
UP000027135 UP000235965 UP000008237 UP000215335 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000005205 UP000078542 UP000078492 UP000007755 UP000002320 UP000053105 UP000008820 UP000075886 UP000085678 UP000075882 UP000075902 UP000075903 UP000007062 UP000075840 UP000076407 UP000075881 UP000075883 UP000075884 UP000076408 UP000075809 UP000078540 UP000076502 UP000075900 UP000075920 UP000225706 UP000075885 UP000092461 UP000000673 UP000092462 UP000095300 UP000192223 UP000075880 UP000007110 UP000037069 UP000092444 UP000095301 UP000001070 UP000001593 UP000001819 UP000007266 UP000009046 UP000268350 UP000019118 UP000030742 UP000053454 UP000230750 UP000192221 UP000002282 UP000007801 UP000091820 UP000053825 UP000000803 UP000000304 UP000001292 UP000008792 UP000245119 UP000054359 UP000008711
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
D3JSV8
A0A2W1BEM8
A0A2A4K859
A0A1E1WRI6
A0A194RFP6
A0A194PPU8
+ More
A0A212FJW8 S4PYB6 H9IZF7 K7J4B1 A0A067RSQ5 A0A2J7RG12 A0A2J7RG05 A0A2J7RG15 E2BKQ2 A0A232F829 E2APE4 A0A2A3E2G3 A0A088ASD0 A0A026VVY8 E9I9Q3 A0A158NTD3 A0A195CLT5 A0A151J9X9 F4W3Y3 B0W3P3 A0A023ERA1 A0A1Q3FEE5 A0A1Y1KMZ7 A0A0N1ITX8 A0A1Y1KMC1 Q17AT2 A0A1B6CL71 A0A1B6C546 A0A1B6H0Y2 A0A1B6INS3 A0A1L8DVF6 A0A182QCF8 A0A1B6M7G7 T1JL47 A0A1S3I7K2 A0A182KRU5 A0A182UDT4 A0A182UVR9 Q7PTD6 A0A182I3Q2 A0A182X6E7 U5EQE9 A0A182JNY2 A0A182MQM4 A0A182NHK8 A0A182XZ98 A0A151X1S2 A0A195B3U0 A0A0T6BAL1 A0A154NZZ5 A0A182RFP1 A0A182WIS2 A0A2B4R6R1 A0A182P3J1 A0A1B0GJ67 W5J9P0 A0A2M4A7C1 A0A0M4MP51 A0A1B0D5F4 A0A2M4BR90 A0A2M3Z4X6 A0A034VEC9 W8BV62 A0A0K8VIP9 A0A1I8PBQ1 A0A1W4WL25 A0A182JFE4 W4ZAA5 A0A0K8TPU2 A0A0L0BPL7 D3TPQ6 T1PC66 B4J8C5 A7SI02 Q28ZX7 D6WDF9 E0VN58 A0A3B0JPG1 N6TKD7 A0A0L8FRD0 A0A2G8LCZ1 A0A1W4WB83 B4P2P3 B3MGU0 A0A1A9X4A0 A0A0J9R963 A0A0L7R403 Q7K519 B4QFB9 B4HRK9 B4LJN3 A0A2T7PF46 A0A087U2C6 B3N8T4
A0A212FJW8 S4PYB6 H9IZF7 K7J4B1 A0A067RSQ5 A0A2J7RG12 A0A2J7RG05 A0A2J7RG15 E2BKQ2 A0A232F829 E2APE4 A0A2A3E2G3 A0A088ASD0 A0A026VVY8 E9I9Q3 A0A158NTD3 A0A195CLT5 A0A151J9X9 F4W3Y3 B0W3P3 A0A023ERA1 A0A1Q3FEE5 A0A1Y1KMZ7 A0A0N1ITX8 A0A1Y1KMC1 Q17AT2 A0A1B6CL71 A0A1B6C546 A0A1B6H0Y2 A0A1B6INS3 A0A1L8DVF6 A0A182QCF8 A0A1B6M7G7 T1JL47 A0A1S3I7K2 A0A182KRU5 A0A182UDT4 A0A182UVR9 Q7PTD6 A0A182I3Q2 A0A182X6E7 U5EQE9 A0A182JNY2 A0A182MQM4 A0A182NHK8 A0A182XZ98 A0A151X1S2 A0A195B3U0 A0A0T6BAL1 A0A154NZZ5 A0A182RFP1 A0A182WIS2 A0A2B4R6R1 A0A182P3J1 A0A1B0GJ67 W5J9P0 A0A2M4A7C1 A0A0M4MP51 A0A1B0D5F4 A0A2M4BR90 A0A2M3Z4X6 A0A034VEC9 W8BV62 A0A0K8VIP9 A0A1I8PBQ1 A0A1W4WL25 A0A182JFE4 W4ZAA5 A0A0K8TPU2 A0A0L0BPL7 D3TPQ6 T1PC66 B4J8C5 A7SI02 Q28ZX7 D6WDF9 E0VN58 A0A3B0JPG1 N6TKD7 A0A0L8FRD0 A0A2G8LCZ1 A0A1W4WB83 B4P2P3 B3MGU0 A0A1A9X4A0 A0A0J9R963 A0A0L7R403 Q7K519 B4QFB9 B4HRK9 B4LJN3 A0A2T7PF46 A0A087U2C6 B3N8T4
PDB
6CES
E-value=6.36236e-136,
Score=1240
Ontologies
PATHWAY
GO
PANTHER
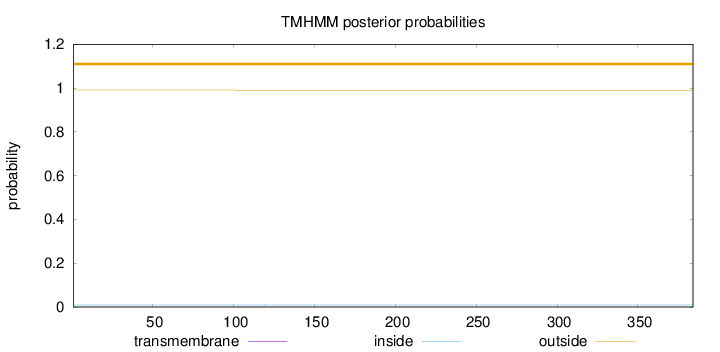
Topology
Length:
384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00491999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00940
outside
1 - 384
Population Genetic Test Statistics
Pi
167.317094
Theta
163.301075
Tajima's D
0.072258
CLR
0.778391
CSRT
0.391230438478076
Interpretation
Uncertain
