Gene
KWMTBOMO02633
Pre Gene Modal
BGIBMGA002641
Annotation
GPI_transamidase_component_PIG-T_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.453
Sequence
CDS
ATGTATGTGTTAGTAAATGATAACCTGTCATCCGAGCACACCTATTTAGCACCTAGGTCTTTGGTGGAAGTGTTGACACGATTCCAAGTTGATGAACTGCATTTCACACTAACCGAGGGGCAGTGGAGGCATAATCATTGGGGTTATCCTGTGCTTGATGCAGCTCCTGGAGCAGAACTATACGCTTGGTTTTCCTCAGATGTGGAAAATGTTGATACACAATGGAAAAAATTAAGTTCCACCCTTGCAGGACTGTTCTGTGCATCTTTAAATTTTATTGAAGATTTCAACACTATAACACCTCAAATGGCATTACAACCGACCGGTGCTATACCACAAAATCCTAATATAACATCACAGTTACGCTATGCTTCTCTGCCTAGAGAAATTGTATGCACAGAAAACTTAACACCATGGAAGAAACTTCTTCCTTGTGAATCTAGTAGAGGGCTTTCAGCATTGTTAAACTCAAGAATGATACACAATACCAATTATCATTCTATTGGAGTTCATACAAGAAAAGTGTGTTCATCTAGTGATTGTTCAGTAGTGAGCCTAGAGATAAAGCAAACGGTAGCTTTGGTTTATGATTCTAAGATTATTCGAACCACTGATTGGTCTTTTACTAAATTATTTGGCCAAGGGTTGCCTGGTGGATGCCCTCTTGCATCAAGTAGCAAAGTGTATGTAGATATCACCACCAATGATACCCAGCCATTCCTTCTATCTCCAGAGCCAGAAAAGGTTCTACTGTCACAGCGAGGAGGCAGTGAAACAAAACTTGCTCTGTACACAATTAAACCTGAAAACAAAATGACTACTATTGGAGCCAAATATATCAGTAAGAAAGATTTGGTTCTTATGAAACTGCCCCCACTTTATTTCAATCGTTATGTATTAGGTTATGGAAAGGAATTTGGAGGTATTGTGACTGAACTGACCAATACTTTTTGGTCCTCCATTGATGTTGTTGTATTGCAGAATGCGCCTTGGTGGTTGCCTATTCAATTAAGCTCTTTAAGAATCAACGGGGAAGCGTCAAGTGAGATGGTGTTAGCTAGGCATTATTCACCTGGGCGTAGCAGACAAAAACCTTATCACTTGGAGTTACTGCTGAGGCTACCACCTAAGTCAGCAACTAAGATAACTATTGATTTTGAATTCGTATTCTTAAAATGGCAAGAGTATCCACCAGATGCTAATCATGGCTTTTATGTTGGGTCAGCAATAATAACTGCTAATTTACCAACAGCAAGAAATTATACTAGTTTACCAGGGACTGCAAGCTTATTTGAATCTTCAATTAATGCATCAAAACCATGGTATCCTGCAGTATTTCGTACTAATGGTGTGATGGTATCATTACCAACACCAGACTTCAGTATGCCATATAATGTGATATGTCTTGCTTGTACAGTTGTGGCCCTTGCTTTTGGACCTCTGCATAATATTTGTACAAAAGAATTAGTTCTCAAAGTTGTTGGTTCTTCTGTTACTTTAAAACAAAAGATTATTAATTTATTTAAAAAGAAAAAGGAGTGA
Protein
MYVLVNDNLSSEHTYLAPRSLVEVLTRFQVDELHFTLTEGQWRHNHWGYPVLDAAPGAELYAWFSSDVENVDTQWKKLSSTLAGLFCASLNFIEDFNTITPQMALQPTGAIPQNPNITSQLRYASLPREIVCTENLTPWKKLLPCESSRGLSALLNSRMIHNTNYHSIGVHTRKVCSSSDCSVVSLEIKQTVALVYDSKIIRTTDWSFTKLFGQGLPGGCPLASSSKVYVDITTNDTQPFLLSPEPEKVLLSQRGGSETKLALYTIKPENKMTTIGAKYISKKDLVLMKLPPLYFNRYVLGYGKEFGGIVTELTNTFWSSIDVVVLQNAPWWLPIQLSSLRINGEASSEMVLARHYSPGRSRQKPYHLELLLRLPPKSATKITIDFEFVFLKWQEYPPDANHGFYVGSAIITANLPTARNYTSLPGTASLFESSINASKPWYPAVFRTNGVMVSLPTPDFSMPYNVICLACTVVALAFGPLHNICTKELVLKVVGSSVTLKQKIINLFKKKKE
Summary
Uniprot
H9IZF6
A0A2W1BIR9
A0A2A4JUH8
A0A194PW08
A0A2H1VA38
A0A194RKY8
+ More
A0A212FJX6 A0A0L7LA77 S4P9E8 D6WNG8 A0A154PII7 A0A1B6KKU4 F4WPY1 A0A1B6FRL9 V5H517 A0A088AUU2 A0A151JCA0 A0A2P8XFW3 A0A2A3E5Y7 A0A232F7G1 A0A151IEA7 K7IN57 A0A151K117 E9ILJ1 A0A151X350 A0A195B5U8 A0A2J7QF02 A0A158NV77 A0A0L7QL77 A0A0C9Q8B3 A0A310SRS7 A0A026WQZ5 A0A067R9L5 E2BTC6 A0A0M8ZZ03 A0A1Y1N965 A0A3L8DN17 U4TZ18 N6TL60 J3JVR4 A0A1B6C6L0 A0A1L8E420 R4WSB2 E0VQA3 A0A2K8JMM0 T1I589 A0A224XLF8 A0A0P4VUU9 A0A1W4XUV2 A0A0A9XUB7 A0A0A9XR98 A0A0K8T9S2 A0A182M716 A0A182YCH1 A0A0A1WEV3 A0A1A9VNM5 A0A034WF00 A0A0K8V2J9 A0A182VFG7 A0A182WRS7 A0A182L021 A0A1B0GBL6 A0A1B0A7P6 A0A182QF72 A0A1B0BBF6 A0A1B0BQS7 A0A1A9XX19 A0A182IBS5 A0A182U1V3 Q7Q8M6 A0A084VU19 A0A182NT74 A0A1S4GYK8 A0A182P4N0 B0XEI9 A0A182IZW0 Q16Q98 A0A1S4FT32 A0A1A9X5E4 A0A2M4BHR6 T1P9B1 A0A0L0CJU9 A0A1Q3FBX1 A0A2M3Z2V1 W8BUW0 A0A182VSH4 A0A2M3ZDZ6 A0A2M4AH00 A0A2M4AHF1 A0A2M4AGU8 W5JG22 A0A182K772 A0A182RWC8 A0A1I8Q3L2 A0A1J1IQG9 A0A182FNB1 T1J0L8 B4L8M6 B4JLL6 B4M1W0
A0A212FJX6 A0A0L7LA77 S4P9E8 D6WNG8 A0A154PII7 A0A1B6KKU4 F4WPY1 A0A1B6FRL9 V5H517 A0A088AUU2 A0A151JCA0 A0A2P8XFW3 A0A2A3E5Y7 A0A232F7G1 A0A151IEA7 K7IN57 A0A151K117 E9ILJ1 A0A151X350 A0A195B5U8 A0A2J7QF02 A0A158NV77 A0A0L7QL77 A0A0C9Q8B3 A0A310SRS7 A0A026WQZ5 A0A067R9L5 E2BTC6 A0A0M8ZZ03 A0A1Y1N965 A0A3L8DN17 U4TZ18 N6TL60 J3JVR4 A0A1B6C6L0 A0A1L8E420 R4WSB2 E0VQA3 A0A2K8JMM0 T1I589 A0A224XLF8 A0A0P4VUU9 A0A1W4XUV2 A0A0A9XUB7 A0A0A9XR98 A0A0K8T9S2 A0A182M716 A0A182YCH1 A0A0A1WEV3 A0A1A9VNM5 A0A034WF00 A0A0K8V2J9 A0A182VFG7 A0A182WRS7 A0A182L021 A0A1B0GBL6 A0A1B0A7P6 A0A182QF72 A0A1B0BBF6 A0A1B0BQS7 A0A1A9XX19 A0A182IBS5 A0A182U1V3 Q7Q8M6 A0A084VU19 A0A182NT74 A0A1S4GYK8 A0A182P4N0 B0XEI9 A0A182IZW0 Q16Q98 A0A1S4FT32 A0A1A9X5E4 A0A2M4BHR6 T1P9B1 A0A0L0CJU9 A0A1Q3FBX1 A0A2M3Z2V1 W8BUW0 A0A182VSH4 A0A2M3ZDZ6 A0A2M4AH00 A0A2M4AHF1 A0A2M4AGU8 W5JG22 A0A182K772 A0A182RWC8 A0A1I8Q3L2 A0A1J1IQG9 A0A182FNB1 T1J0L8 B4L8M6 B4JLL6 B4M1W0
Pubmed
19121390
28756777
26354079
22118469
26227816
23622113
+ More
18362917 19820115 21719571 29403074 28648823 20075255 21282665 21347285 24508170 24845553 20798317 28004739 30249741 23537049 22516182 23691247 20566863 27129103 25401762 25244985 25830018 25348373 20966253 12364791 24438588 17510324 25315136 26108605 24495485 20920257 23761445 17994087
18362917 19820115 21719571 29403074 28648823 20075255 21282665 21347285 24508170 24845553 20798317 28004739 30249741 23537049 22516182 23691247 20566863 27129103 25401762 25244985 25830018 25348373 20966253 12364791 24438588 17510324 25315136 26108605 24495485 20920257 23761445 17994087
EMBL
BABH01014142
KZ150098
PZC73584.1
NWSH01000598
PCG75426.1
KQ459589
+ More
KPI97512.1 ODYU01001466 SOQ37699.1 KQ460207 KPJ16641.1 AGBW02008206 OWR54026.1 JTDY01002026 KOB72299.1 GAIX01005521 JAA87039.1 KQ971343 EFA03225.1 KQ434926 KZC11686.1 GEBQ01027900 JAT12077.1 GL888256 EGI63749.1 GECZ01016914 JAS52855.1 GALX01000568 JAB67898.1 KQ979060 KYN23171.1 PYGN01002287 PSN30900.1 KZ288356 PBC27173.1 NNAY01000810 OXU26388.1 KQ977901 KYM98897.1 KQ981189 KYN45250.1 GL764087 EFZ18562.1 KQ982577 KYQ54638.1 KQ976595 KYM79645.1 NEVH01015305 PNF27160.1 ADTU01026947 ADTU01026948 ADTU01026949 ADTU01026950 ADTU01026951 KQ414934 KOC59345.1 GBYB01010468 JAG80235.1 KQ759866 OAD62487.1 KK107128 EZA58388.1 KK852655 KDR19304.1 GL450389 EFN81047.1 KQ435794 KOX73990.1 GEZM01009645 JAV94433.1 QOIP01000006 RLU21845.1 KB631868 ERL86829.1 APGK01032729 KB740848 ENN78703.1 BT127332 AEE62294.1 GEDC01028191 JAS09107.1 GFDF01000812 JAV13272.1 AK417587 BAN20802.1 DS235408 EEB15559.1 KY031140 ATU82891.1 ACPB03008476 GFTR01007443 JAW08983.1 GDKW01002007 JAI54588.1 GBHO01021111 JAG22493.1 GBHO01021110 JAG22494.1 GBRD01003537 JAG62284.1 AXCM01002684 GBXI01017081 JAC97210.1 GAKP01005683 JAC53269.1 GDHF01019162 JAI33152.1 CCAG010001819 AXCN02000727 JXJN01011422 JXJN01018767 APCN01000732 AAAB01008944 EAA10074.3 ATLV01016628 KE525098 KFB41463.1 DS232835 EDS26016.1 CH477756 EAT36563.1 GGFJ01003441 MBW52582.1 KA645317 AFP59946.1 JRES01000292 KNC32683.1 GFDL01009971 JAV25074.1 GGFM01002089 MBW22840.1 GAMC01003593 JAC02963.1 GGFM01006005 MBW26756.1 GGFK01006740 MBW40061.1 GGFK01006717 MBW40038.1 GGFK01006694 MBW40015.1 ADMH02001323 ETN63016.1 CVRI01000056 CRL01788.1 JH431738 CH933815 EDW08001.1 CH916370 EDW00469.1 CH940651 EDW65664.1
KPI97512.1 ODYU01001466 SOQ37699.1 KQ460207 KPJ16641.1 AGBW02008206 OWR54026.1 JTDY01002026 KOB72299.1 GAIX01005521 JAA87039.1 KQ971343 EFA03225.1 KQ434926 KZC11686.1 GEBQ01027900 JAT12077.1 GL888256 EGI63749.1 GECZ01016914 JAS52855.1 GALX01000568 JAB67898.1 KQ979060 KYN23171.1 PYGN01002287 PSN30900.1 KZ288356 PBC27173.1 NNAY01000810 OXU26388.1 KQ977901 KYM98897.1 KQ981189 KYN45250.1 GL764087 EFZ18562.1 KQ982577 KYQ54638.1 KQ976595 KYM79645.1 NEVH01015305 PNF27160.1 ADTU01026947 ADTU01026948 ADTU01026949 ADTU01026950 ADTU01026951 KQ414934 KOC59345.1 GBYB01010468 JAG80235.1 KQ759866 OAD62487.1 KK107128 EZA58388.1 KK852655 KDR19304.1 GL450389 EFN81047.1 KQ435794 KOX73990.1 GEZM01009645 JAV94433.1 QOIP01000006 RLU21845.1 KB631868 ERL86829.1 APGK01032729 KB740848 ENN78703.1 BT127332 AEE62294.1 GEDC01028191 JAS09107.1 GFDF01000812 JAV13272.1 AK417587 BAN20802.1 DS235408 EEB15559.1 KY031140 ATU82891.1 ACPB03008476 GFTR01007443 JAW08983.1 GDKW01002007 JAI54588.1 GBHO01021111 JAG22493.1 GBHO01021110 JAG22494.1 GBRD01003537 JAG62284.1 AXCM01002684 GBXI01017081 JAC97210.1 GAKP01005683 JAC53269.1 GDHF01019162 JAI33152.1 CCAG010001819 AXCN02000727 JXJN01011422 JXJN01018767 APCN01000732 AAAB01008944 EAA10074.3 ATLV01016628 KE525098 KFB41463.1 DS232835 EDS26016.1 CH477756 EAT36563.1 GGFJ01003441 MBW52582.1 KA645317 AFP59946.1 JRES01000292 KNC32683.1 GFDL01009971 JAV25074.1 GGFM01002089 MBW22840.1 GAMC01003593 JAC02963.1 GGFM01006005 MBW26756.1 GGFK01006740 MBW40061.1 GGFK01006717 MBW40038.1 GGFK01006694 MBW40015.1 ADMH02001323 ETN63016.1 CVRI01000056 CRL01788.1 JH431738 CH933815 EDW08001.1 CH916370 EDW00469.1 CH940651 EDW65664.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000076502 UP000007755 UP000005203 UP000078492 UP000245037 UP000242457 UP000215335 UP000078542 UP000002358 UP000078541 UP000075809 UP000078540 UP000235965 UP000005205 UP000053825 UP000053097 UP000027135 UP000008237 UP000053105 UP000279307 UP000030742 UP000019118 UP000009046 UP000015103 UP000192223 UP000075883 UP000076408 UP000078200 UP000075903 UP000076407 UP000075882 UP000092444 UP000092445 UP000075886 UP000092460 UP000092443 UP000075840 UP000075902 UP000007062 UP000030765 UP000075884 UP000075885 UP000002320 UP000075880 UP000008820 UP000091820 UP000095301 UP000037069 UP000075920 UP000000673 UP000075881 UP000075900 UP000095300 UP000183832 UP000069272 UP000009192 UP000001070 UP000008792
UP000007266 UP000076502 UP000007755 UP000005203 UP000078492 UP000245037 UP000242457 UP000215335 UP000078542 UP000002358 UP000078541 UP000075809 UP000078540 UP000235965 UP000005205 UP000053825 UP000053097 UP000027135 UP000008237 UP000053105 UP000279307 UP000030742 UP000019118 UP000009046 UP000015103 UP000192223 UP000075883 UP000076408 UP000078200 UP000075903 UP000076407 UP000075882 UP000092444 UP000092445 UP000075886 UP000092460 UP000092443 UP000075840 UP000075902 UP000007062 UP000030765 UP000075884 UP000075885 UP000002320 UP000075880 UP000008820 UP000091820 UP000095301 UP000037069 UP000075920 UP000000673 UP000075881 UP000075900 UP000095300 UP000183832 UP000069272 UP000009192 UP000001070 UP000008792
PRIDE
Interpro
SUPFAM
SSF54913
SSF54913
Gene 3D
ProteinModelPortal
H9IZF6
A0A2W1BIR9
A0A2A4JUH8
A0A194PW08
A0A2H1VA38
A0A194RKY8
+ More
A0A212FJX6 A0A0L7LA77 S4P9E8 D6WNG8 A0A154PII7 A0A1B6KKU4 F4WPY1 A0A1B6FRL9 V5H517 A0A088AUU2 A0A151JCA0 A0A2P8XFW3 A0A2A3E5Y7 A0A232F7G1 A0A151IEA7 K7IN57 A0A151K117 E9ILJ1 A0A151X350 A0A195B5U8 A0A2J7QF02 A0A158NV77 A0A0L7QL77 A0A0C9Q8B3 A0A310SRS7 A0A026WQZ5 A0A067R9L5 E2BTC6 A0A0M8ZZ03 A0A1Y1N965 A0A3L8DN17 U4TZ18 N6TL60 J3JVR4 A0A1B6C6L0 A0A1L8E420 R4WSB2 E0VQA3 A0A2K8JMM0 T1I589 A0A224XLF8 A0A0P4VUU9 A0A1W4XUV2 A0A0A9XUB7 A0A0A9XR98 A0A0K8T9S2 A0A182M716 A0A182YCH1 A0A0A1WEV3 A0A1A9VNM5 A0A034WF00 A0A0K8V2J9 A0A182VFG7 A0A182WRS7 A0A182L021 A0A1B0GBL6 A0A1B0A7P6 A0A182QF72 A0A1B0BBF6 A0A1B0BQS7 A0A1A9XX19 A0A182IBS5 A0A182U1V3 Q7Q8M6 A0A084VU19 A0A182NT74 A0A1S4GYK8 A0A182P4N0 B0XEI9 A0A182IZW0 Q16Q98 A0A1S4FT32 A0A1A9X5E4 A0A2M4BHR6 T1P9B1 A0A0L0CJU9 A0A1Q3FBX1 A0A2M3Z2V1 W8BUW0 A0A182VSH4 A0A2M3ZDZ6 A0A2M4AH00 A0A2M4AHF1 A0A2M4AGU8 W5JG22 A0A182K772 A0A182RWC8 A0A1I8Q3L2 A0A1J1IQG9 A0A182FNB1 T1J0L8 B4L8M6 B4JLL6 B4M1W0
A0A212FJX6 A0A0L7LA77 S4P9E8 D6WNG8 A0A154PII7 A0A1B6KKU4 F4WPY1 A0A1B6FRL9 V5H517 A0A088AUU2 A0A151JCA0 A0A2P8XFW3 A0A2A3E5Y7 A0A232F7G1 A0A151IEA7 K7IN57 A0A151K117 E9ILJ1 A0A151X350 A0A195B5U8 A0A2J7QF02 A0A158NV77 A0A0L7QL77 A0A0C9Q8B3 A0A310SRS7 A0A026WQZ5 A0A067R9L5 E2BTC6 A0A0M8ZZ03 A0A1Y1N965 A0A3L8DN17 U4TZ18 N6TL60 J3JVR4 A0A1B6C6L0 A0A1L8E420 R4WSB2 E0VQA3 A0A2K8JMM0 T1I589 A0A224XLF8 A0A0P4VUU9 A0A1W4XUV2 A0A0A9XUB7 A0A0A9XR98 A0A0K8T9S2 A0A182M716 A0A182YCH1 A0A0A1WEV3 A0A1A9VNM5 A0A034WF00 A0A0K8V2J9 A0A182VFG7 A0A182WRS7 A0A182L021 A0A1B0GBL6 A0A1B0A7P6 A0A182QF72 A0A1B0BBF6 A0A1B0BQS7 A0A1A9XX19 A0A182IBS5 A0A182U1V3 Q7Q8M6 A0A084VU19 A0A182NT74 A0A1S4GYK8 A0A182P4N0 B0XEI9 A0A182IZW0 Q16Q98 A0A1S4FT32 A0A1A9X5E4 A0A2M4BHR6 T1P9B1 A0A0L0CJU9 A0A1Q3FBX1 A0A2M3Z2V1 W8BUW0 A0A182VSH4 A0A2M3ZDZ6 A0A2M4AH00 A0A2M4AHF1 A0A2M4AGU8 W5JG22 A0A182K772 A0A182RWC8 A0A1I8Q3L2 A0A1J1IQG9 A0A182FNB1 T1J0L8 B4L8M6 B4JLL6 B4M1W0
Ontologies
PATHWAY
GO
PANTHER
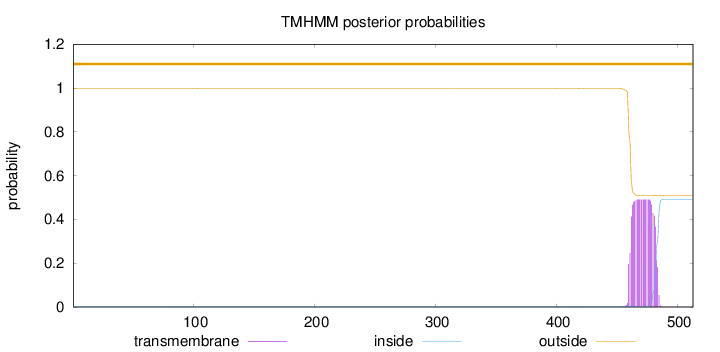
Topology
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.86686
Exp number, first 60 AAs:
0.0003
Total prob of N-in:
0.00327
outside
1 - 513
Population Genetic Test Statistics
Pi
180.251915
Theta
164.766156
Tajima's D
0.228413
CLR
0.014419
CSRT
0.436578171091445
Interpretation
Uncertain
