Gene
KWMTBOMO02631
Annotation
PREDICTED:_major_facilitator_superfamily_domain-containing_protein_12-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.971
Sequence
CDS
ATGGATAACGAATTTATAGATATATCGACAAGTCTCAAACTACAACTGGGCTATGGAATTGGTCACGTACTTAATGATGTTTGTGCCAGTTTGTGGTTTACATATTTTCTAGTATATTTTAAACTAGTACTAGAGTTCAGTAATGTGCAGTCTGGTACACTTATGTTAGTAGGTCAAATGGTTGATGCTTTAGCCACTCCTTTTATTGGTTATCACTCGGATCATACAGACAATTTCTGGAGTGCAAAATATGGAAGACGGAAGCTCTGGCATTTGTTTGGTACAATCTGTGTTGTGACCTCATTTCCATTTATCTTTTCTGAATGTGTTGGATGTTCATTGACACACAGATGGGCTCAAATGTACTATTTTGCTGCTTTTATTGTCATATTCCAAATTGGATGGGCAGCAGTGCAAATTTCGCATTTGAGCCTAATACCTGAATTAGCACAAGATGATCACACAAGAACACACTTGACCGCTATCAGATATGCATTCACTGTGTTCTCTAATTTGTTGGTGTACATTGCTACGTGGATAATATTGCATATTACAGGAGAATGTGACAAAAAGCAAGTAGGGCCATCTGACAGTTGGAAATTCAGGCACATAGTTTTTATAGTTATGGGTGTTGGCACTATAGCCTCACTATTGTTTCATTTAAGTGTCAAAGAAACACAAAATGCACGGAGTCGCCAGGTTTTAATAGATGCTGAAGAAAGCGGTGCTCACAACCTTTTCATTCGTAAACCACTTTTATATCAGGTAGCCGGAGTATATATGAGTACAAGGCTAGTAGTTAACTTATCGCAGGTGCTCATACCCTTATATCTTCACCTTACTTTAGGCCTAGCTGCGCGCGCTCTAGCTGCTATACCACTCGCACTGTACATTGGTAGTTTAACAGCAGCAGGATTTCAGAGGTTGGCTCCGAGATCATTTTCAAGAAAAATAAACTACTTTTTTGGTTCCACATGTGCAATGGCTGGTTTTGTATGGATATATTTTGGATCAGATCACAACTATAAGGAGTATTTCATTTATTTGGTCGCATTACTTATAGGTTTTGGTGGTGCAGTTATGTTAGTAACGAGTCTTGCGCTAACAGCAGACTTGATAGGCGAAAGAACCGAGGCTTCTGCTTTTGTATATGGATTAATGAGTTTTAGTGACAAATTGTCGTGTGGACTAGCCATTGCTTTAGTGCAAACATATGCCGACGGAGCTGAATCAACATACTATCGGAATGCTTTGGTGTGGGTGTGCGGAGTTGCAACGATAACTGGCCTCATCTTTACATTATTATTACCTAAATTTCAAACAGATGCCCTTGTTTTGAATGATGGTGCGGCAGACAGTAACACAGAAGATGCGACGAGTGCACCAATAGATAATATATGA
Protein
MDNEFIDISTSLKLQLGYGIGHVLNDVCASLWFTYFLVYFKLVLEFSNVQSGTLMLVGQMVDALATPFIGYHSDHTDNFWSAKYGRRKLWHLFGTICVVTSFPFIFSECVGCSLTHRWAQMYYFAAFIVIFQIGWAAVQISHLSLIPELAQDDHTRTHLTAIRYAFTVFSNLLVYIATWIILHITGECDKKQVGPSDSWKFRHIVFIVMGVGTIASLLFHLSVKETQNARSRQVLIDAEESGAHNLFIRKPLLYQVAGVYMSTRLVVNLSQVLIPLYLHLTLGLAARALAAIPLALYIGSLTAAGFQRLAPRSFSRKINYFFGSTCAMAGFVWIYFGSDHNYKEYFIYLVALLIGFGGAVMLVTSLALTADLIGERTEASAFVYGLMSFSDKLSCGLAIALVQTYADGAESTYYRNALVWVCGVATITGLIFTLLLPKFQTDALVLNDGAADSNTEDATSAPIDNI
Summary
Uniprot
A0A2H1VA33
A0A2A4JTR3
A0A2W1CHU9
A0A194PWW8
A0A212FJU5
A0A1Y1LD93
+ More
A0A1W4XDI7 D6W847 A0A2J7PGJ2 A0A139WPG3 K7IS83 A0A1W4XMR4 A0A232ERE3 A0A067R5D6 A0A088AJC7 E2BGG8 E2AJ53 A0A195BKU0 A0A158P455 A0A151JVP9 A0A151J9B2 A0A195CNW3 A0A0L7RBV1 A0A0C9QWP9 A0A151X0H6 N6TV38 U4U133 E0VQM9 A0A154PGK0 T1JDD2 A0A087V0H2 A0A0P4WDL5 A0A2R5L9G5 A0A1Z5L4U1 U4U742 A0A1B6E3G3 J3JWL9 N6TG11 A0A1B6DY20 A0A1Y1JXQ6 A0A1Y1JUN1 V5IFC7 A0A1B6IVT6 A0A1B6HVZ9 E9FVJ5 A0A2B4RFV9 A0A0P5A7W7 A0A0P5NTE2 A0A0P5JVG7 A0A0P5RMK7 A0A0P5V7Q2 A0A0P5AB09 A0A0N8A344 A0A0P5VXK5 A0A0P5N487 A0A0P5AWT0 A0A0P5IX88 A0A0P6HIP6 L7M8D3 A0A0P4WNU2 A0A0P4Z4M9 A0A131XCD3 V4B2M1 A0A1E1X3L5 A0A1B6M148 A0A131YXE6 A0A2R7WKZ4 A0A224Z9C2 A7SNU2 E9J1Q3 A0A1B6MRX0 V5H9F2 A0A1W0WRE6 A0A3M6U119 A0A210PMX4 G3W051 A0A3B4BXG8 I3JY70 G1PQY8 A0A2I4B9N9 A0A2I4D7R3 A0A0P4VN71 F1MZJ3 A0A2D0QVM8 A0A3P9CUQ6 W5PS21 W5LZ76 Q1LWK8 T1I071 K9J160 H3B654 F1R9L4 A0A2G8L5E6 M4AWS7 A0A3B3T7E1 R7UXX9 A0A224XQ27 A0A023F164 A0A369RST5
A0A1W4XDI7 D6W847 A0A2J7PGJ2 A0A139WPG3 K7IS83 A0A1W4XMR4 A0A232ERE3 A0A067R5D6 A0A088AJC7 E2BGG8 E2AJ53 A0A195BKU0 A0A158P455 A0A151JVP9 A0A151J9B2 A0A195CNW3 A0A0L7RBV1 A0A0C9QWP9 A0A151X0H6 N6TV38 U4U133 E0VQM9 A0A154PGK0 T1JDD2 A0A087V0H2 A0A0P4WDL5 A0A2R5L9G5 A0A1Z5L4U1 U4U742 A0A1B6E3G3 J3JWL9 N6TG11 A0A1B6DY20 A0A1Y1JXQ6 A0A1Y1JUN1 V5IFC7 A0A1B6IVT6 A0A1B6HVZ9 E9FVJ5 A0A2B4RFV9 A0A0P5A7W7 A0A0P5NTE2 A0A0P5JVG7 A0A0P5RMK7 A0A0P5V7Q2 A0A0P5AB09 A0A0N8A344 A0A0P5VXK5 A0A0P5N487 A0A0P5AWT0 A0A0P5IX88 A0A0P6HIP6 L7M8D3 A0A0P4WNU2 A0A0P4Z4M9 A0A131XCD3 V4B2M1 A0A1E1X3L5 A0A1B6M148 A0A131YXE6 A0A2R7WKZ4 A0A224Z9C2 A7SNU2 E9J1Q3 A0A1B6MRX0 V5H9F2 A0A1W0WRE6 A0A3M6U119 A0A210PMX4 G3W051 A0A3B4BXG8 I3JY70 G1PQY8 A0A2I4B9N9 A0A2I4D7R3 A0A0P4VN71 F1MZJ3 A0A2D0QVM8 A0A3P9CUQ6 W5PS21 W5LZ76 Q1LWK8 T1I071 K9J160 H3B654 F1R9L4 A0A2G8L5E6 M4AWS7 A0A3B3T7E1 R7UXX9 A0A224XQ27 A0A023F164 A0A369RST5
Pubmed
28756777
26354079
22118469
28004739
18362917
19820115
+ More
20075255 28648823 24845553 20798317 21347285 23537049 20566863 28528879 22516182 25765539 21292972 25576852 28049606 23254933 28503490 26830274 28797301 17615350 21282665 30382153 28812685 21709235 25186727 21993624 27129103 19393038 20809919 23594743 9215903 29023486 23542700 29240929 25474469 30042472
20075255 28648823 24845553 20798317 21347285 23537049 20566863 28528879 22516182 25765539 21292972 25576852 28049606 23254933 28503490 26830274 28797301 17615350 21282665 30382153 28812685 21709235 25186727 21993624 27129103 19393038 20809919 23594743 9215903 29023486 23542700 29240929 25474469 30042472
EMBL
ODYU01001466
SOQ37698.1
NWSH01000598
PCG75425.1
KZ150098
PZC73585.1
+ More
KQ459589 KPI97513.1 AGBW02008206 OWR54027.1 GEZM01058973 JAV71594.1 KQ971307 EFA10999.1 NEVH01025212 PNF15452.1 KYB29834.1 NNAY01002629 OXU20901.1 KK852954 KDR13311.1 GL448181 EFN85217.1 GL439967 EFN66493.1 KQ976453 KYM85340.1 ADTU01008610 KQ981701 KYN37495.1 KQ979480 KYN21281.1 KQ977513 KYN02167.1 KQ414617 KOC68329.1 GBYB01008159 JAG77926.1 KQ982612 KYQ53868.1 APGK01050965 APGK01050966 KB741180 ENN73125.1 KB631768 ERL85998.1 DS235430 EEB15685.1 KQ434899 KZC10942.1 JH432104 KK122592 KFM83111.1 GDRN01071210 GDRN01071209 JAI63734.1 GGLE01001989 MBY06115.1 GFJQ02004591 JAW02379.1 KB631815 ERL86416.1 GEDC01004847 JAS32451.1 BT127637 AEE62599.1 APGK01029196 KB740727 ENN79329.1 GEDC01006737 JAS30561.1 GEZM01100145 JAV52991.1 GEZM01100144 JAV52992.1 GANP01008667 JAB75801.1 GECU01032125 GECU01016756 JAS75581.1 JAS90950.1 GECU01035747 GECU01031281 GECU01028890 GECU01004973 JAS71959.1 JAS76425.1 JAS78816.1 JAT02734.1 GL732525 EFX89093.1 LSMT01000485 PFX17254.1 GDIP01207236 JAJ16166.1 GDIQ01148232 JAL03494.1 GDIQ01193458 JAK58267.1 GDIQ01117493 JAL34233.1 GDIP01103227 JAM00488.1 GDIP01201627 JAJ21775.1 GDIP01187185 GDIP01108483 LRGB01002864 JAJ36217.1 KZS05761.1 GDIP01094649 JAM09066.1 GDIQ01149839 JAL01887.1 GDIP01207237 JAJ16165.1 GDIQ01209319 JAK42406.1 GDIQ01018585 JAN76152.1 GACK01004932 JAA60102.1 GDIP01254223 JAI69178.1 GDIP01222376 JAJ01026.1 GEFH01003718 JAP64863.1 KB200701 ESP00677.1 GFAC01005329 JAT93859.1 GEBQ01029133 GEBQ01010357 JAT10844.1 JAT29620.1 GEDV01004920 JAP83637.1 KK855011 PTY20278.1 GFPF01012118 MAA23264.1 DS469724 EDO34623.1 GL767674 EFZ13257.1 GEBQ01001321 JAT38656.1 GANP01013171 JAB71297.1 MTYJ01000056 OQV17781.1 RCHS01002420 RMX47405.1 NEDP02005580 OWF37776.1 AEFK01069492 AEFK01069493 AEFK01069494 AEFK01069495 AERX01055838 AERX01055839 AAPE02052840 GDKW01002662 JAI53933.1 AMGL01091633 AHAT01011258 BX548024 ACPB03023664 GABZ01005886 JAA47639.1 AFYH01099213 AFYH01099214 AFYH01099215 AFYH01099216 MRZV01000213 PIK55499.1 AMQN01006812 KB299181 ELU08281.1 GFTR01006327 JAW10099.1 GBBI01003951 JAC14761.1 NOWV01000217 RDD37755.1
KQ459589 KPI97513.1 AGBW02008206 OWR54027.1 GEZM01058973 JAV71594.1 KQ971307 EFA10999.1 NEVH01025212 PNF15452.1 KYB29834.1 NNAY01002629 OXU20901.1 KK852954 KDR13311.1 GL448181 EFN85217.1 GL439967 EFN66493.1 KQ976453 KYM85340.1 ADTU01008610 KQ981701 KYN37495.1 KQ979480 KYN21281.1 KQ977513 KYN02167.1 KQ414617 KOC68329.1 GBYB01008159 JAG77926.1 KQ982612 KYQ53868.1 APGK01050965 APGK01050966 KB741180 ENN73125.1 KB631768 ERL85998.1 DS235430 EEB15685.1 KQ434899 KZC10942.1 JH432104 KK122592 KFM83111.1 GDRN01071210 GDRN01071209 JAI63734.1 GGLE01001989 MBY06115.1 GFJQ02004591 JAW02379.1 KB631815 ERL86416.1 GEDC01004847 JAS32451.1 BT127637 AEE62599.1 APGK01029196 KB740727 ENN79329.1 GEDC01006737 JAS30561.1 GEZM01100145 JAV52991.1 GEZM01100144 JAV52992.1 GANP01008667 JAB75801.1 GECU01032125 GECU01016756 JAS75581.1 JAS90950.1 GECU01035747 GECU01031281 GECU01028890 GECU01004973 JAS71959.1 JAS76425.1 JAS78816.1 JAT02734.1 GL732525 EFX89093.1 LSMT01000485 PFX17254.1 GDIP01207236 JAJ16166.1 GDIQ01148232 JAL03494.1 GDIQ01193458 JAK58267.1 GDIQ01117493 JAL34233.1 GDIP01103227 JAM00488.1 GDIP01201627 JAJ21775.1 GDIP01187185 GDIP01108483 LRGB01002864 JAJ36217.1 KZS05761.1 GDIP01094649 JAM09066.1 GDIQ01149839 JAL01887.1 GDIP01207237 JAJ16165.1 GDIQ01209319 JAK42406.1 GDIQ01018585 JAN76152.1 GACK01004932 JAA60102.1 GDIP01254223 JAI69178.1 GDIP01222376 JAJ01026.1 GEFH01003718 JAP64863.1 KB200701 ESP00677.1 GFAC01005329 JAT93859.1 GEBQ01029133 GEBQ01010357 JAT10844.1 JAT29620.1 GEDV01004920 JAP83637.1 KK855011 PTY20278.1 GFPF01012118 MAA23264.1 DS469724 EDO34623.1 GL767674 EFZ13257.1 GEBQ01001321 JAT38656.1 GANP01013171 JAB71297.1 MTYJ01000056 OQV17781.1 RCHS01002420 RMX47405.1 NEDP02005580 OWF37776.1 AEFK01069492 AEFK01069493 AEFK01069494 AEFK01069495 AERX01055838 AERX01055839 AAPE02052840 GDKW01002662 JAI53933.1 AMGL01091633 AHAT01011258 BX548024 ACPB03023664 GABZ01005886 JAA47639.1 AFYH01099213 AFYH01099214 AFYH01099215 AFYH01099216 MRZV01000213 PIK55499.1 AMQN01006812 KB299181 ELU08281.1 GFTR01006327 JAW10099.1 GBBI01003951 JAC14761.1 NOWV01000217 RDD37755.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000192223
UP000007266
UP000235965
+ More
UP000002358 UP000215335 UP000027135 UP000005203 UP000008237 UP000000311 UP000078540 UP000005205 UP000078541 UP000078492 UP000078542 UP000053825 UP000075809 UP000019118 UP000030742 UP000009046 UP000076502 UP000054359 UP000000305 UP000225706 UP000076858 UP000030746 UP000001593 UP000275408 UP000242188 UP000007648 UP000261440 UP000005207 UP000001074 UP000192220 UP000009136 UP000221080 UP000265160 UP000002356 UP000018468 UP000000437 UP000015103 UP000008672 UP000230750 UP000002852 UP000261540 UP000014760 UP000253843
UP000002358 UP000215335 UP000027135 UP000005203 UP000008237 UP000000311 UP000078540 UP000005205 UP000078541 UP000078492 UP000078542 UP000053825 UP000075809 UP000019118 UP000030742 UP000009046 UP000076502 UP000054359 UP000000305 UP000225706 UP000076858 UP000030746 UP000001593 UP000275408 UP000242188 UP000007648 UP000261440 UP000005207 UP000001074 UP000192220 UP000009136 UP000221080 UP000265160 UP000002356 UP000018468 UP000000437 UP000015103 UP000008672 UP000230750 UP000002852 UP000261540 UP000014760 UP000253843
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1VA33
A0A2A4JTR3
A0A2W1CHU9
A0A194PWW8
A0A212FJU5
A0A1Y1LD93
+ More
A0A1W4XDI7 D6W847 A0A2J7PGJ2 A0A139WPG3 K7IS83 A0A1W4XMR4 A0A232ERE3 A0A067R5D6 A0A088AJC7 E2BGG8 E2AJ53 A0A195BKU0 A0A158P455 A0A151JVP9 A0A151J9B2 A0A195CNW3 A0A0L7RBV1 A0A0C9QWP9 A0A151X0H6 N6TV38 U4U133 E0VQM9 A0A154PGK0 T1JDD2 A0A087V0H2 A0A0P4WDL5 A0A2R5L9G5 A0A1Z5L4U1 U4U742 A0A1B6E3G3 J3JWL9 N6TG11 A0A1B6DY20 A0A1Y1JXQ6 A0A1Y1JUN1 V5IFC7 A0A1B6IVT6 A0A1B6HVZ9 E9FVJ5 A0A2B4RFV9 A0A0P5A7W7 A0A0P5NTE2 A0A0P5JVG7 A0A0P5RMK7 A0A0P5V7Q2 A0A0P5AB09 A0A0N8A344 A0A0P5VXK5 A0A0P5N487 A0A0P5AWT0 A0A0P5IX88 A0A0P6HIP6 L7M8D3 A0A0P4WNU2 A0A0P4Z4M9 A0A131XCD3 V4B2M1 A0A1E1X3L5 A0A1B6M148 A0A131YXE6 A0A2R7WKZ4 A0A224Z9C2 A7SNU2 E9J1Q3 A0A1B6MRX0 V5H9F2 A0A1W0WRE6 A0A3M6U119 A0A210PMX4 G3W051 A0A3B4BXG8 I3JY70 G1PQY8 A0A2I4B9N9 A0A2I4D7R3 A0A0P4VN71 F1MZJ3 A0A2D0QVM8 A0A3P9CUQ6 W5PS21 W5LZ76 Q1LWK8 T1I071 K9J160 H3B654 F1R9L4 A0A2G8L5E6 M4AWS7 A0A3B3T7E1 R7UXX9 A0A224XQ27 A0A023F164 A0A369RST5
A0A1W4XDI7 D6W847 A0A2J7PGJ2 A0A139WPG3 K7IS83 A0A1W4XMR4 A0A232ERE3 A0A067R5D6 A0A088AJC7 E2BGG8 E2AJ53 A0A195BKU0 A0A158P455 A0A151JVP9 A0A151J9B2 A0A195CNW3 A0A0L7RBV1 A0A0C9QWP9 A0A151X0H6 N6TV38 U4U133 E0VQM9 A0A154PGK0 T1JDD2 A0A087V0H2 A0A0P4WDL5 A0A2R5L9G5 A0A1Z5L4U1 U4U742 A0A1B6E3G3 J3JWL9 N6TG11 A0A1B6DY20 A0A1Y1JXQ6 A0A1Y1JUN1 V5IFC7 A0A1B6IVT6 A0A1B6HVZ9 E9FVJ5 A0A2B4RFV9 A0A0P5A7W7 A0A0P5NTE2 A0A0P5JVG7 A0A0P5RMK7 A0A0P5V7Q2 A0A0P5AB09 A0A0N8A344 A0A0P5VXK5 A0A0P5N487 A0A0P5AWT0 A0A0P5IX88 A0A0P6HIP6 L7M8D3 A0A0P4WNU2 A0A0P4Z4M9 A0A131XCD3 V4B2M1 A0A1E1X3L5 A0A1B6M148 A0A131YXE6 A0A2R7WKZ4 A0A224Z9C2 A7SNU2 E9J1Q3 A0A1B6MRX0 V5H9F2 A0A1W0WRE6 A0A3M6U119 A0A210PMX4 G3W051 A0A3B4BXG8 I3JY70 G1PQY8 A0A2I4B9N9 A0A2I4D7R3 A0A0P4VN71 F1MZJ3 A0A2D0QVM8 A0A3P9CUQ6 W5PS21 W5LZ76 Q1LWK8 T1I071 K9J160 H3B654 F1R9L4 A0A2G8L5E6 M4AWS7 A0A3B3T7E1 R7UXX9 A0A224XQ27 A0A023F164 A0A369RST5
PDB
4M64
E-value=8.84422e-07,
Score=127
Ontologies
KEGG
PANTHER
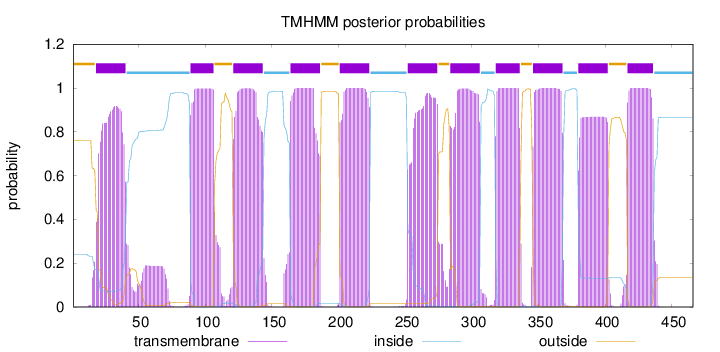
Topology
Length:
466
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
237.85603
Exp number, first 60 AAs:
22.0711
Total prob of N-in:
0.24031
POSSIBLE N-term signal
sequence
outside
1 - 17
TMhelix
18 - 40
inside
41 - 88
TMhelix
89 - 106
outside
107 - 120
TMhelix
121 - 143
inside
144 - 163
TMhelix
164 - 186
outside
187 - 200
TMhelix
201 - 223
inside
224 - 251
TMhelix
252 - 274
outside
275 - 283
TMhelix
284 - 306
inside
307 - 317
TMhelix
318 - 336
outside
337 - 345
TMhelix
346 - 368
inside
369 - 379
TMhelix
380 - 402
outside
403 - 416
TMhelix
417 - 436
inside
437 - 466
Population Genetic Test Statistics
Pi
248.781285
Theta
181.203372
Tajima's D
1.188795
CLR
0.053259
CSRT
0.707714614269287
Interpretation
Uncertain
