Gene
KWMTBOMO02630 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002640
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_[Bombyx_mori]
Full name
Ubiquitin carboxyl-terminal hydrolase
Alternative Name
Ubiquitin thioesterase
Location in the cell
Cytoplasmic Reliability : 2.948
Sequence
CDS
ATGGCGACCGAAACTTTAGTCCCTCTTGAATCCAATCCCGATGTCCTGAACAAGTTTTTGCAAAAGCTCGGCGTACCAAACAAATGGAATATTGTAGATGTCATGGGACTTGATCCTGAAACGCTTTCGTGGGTACCTCGCCCTGTGCTTTCTGTAATGCTACTTTTCCCAATTTCTGATGCTTACGAAAATCACAAAAAAACTGAAGAAAATGAGATTCTGTCCAAAGGGCAAGAAGTTTCTGGAAATATTTTCTATATGAAACAAAATATCAGCAATGCATGTGGGACTATAGCTCTTGTACATAGTGTGGCCAACAATACGGACATAATTGAATTATCTGATGGTCATATGCAGAAATTTTTAAATGAAGCTAAAGGACTTGATGCTACTGCTCGTGGTAAATTGTTGGAAAAGTCTGAAGGCATCATTAATGCTCACAAGGAATTGGCTCAAGAAGGCCAAACAAATACACCAAGTGCTGAAGATCCCGTCAACCATCATTTTATCAGCTTTGTCCAGAAAGATGGAGCATTGTATGAGTTAGATGGCCGTAAAGCTTTTCCTGTCAATCATGGCCCAACTTCACAGGAAACTTTGTTGGAAGATGCAGCAAAAATTTGCAAAGAATTTATGGCTCGCGATCCGAATGAAGTACGCTTTACAGTAATTGCTCTAGTAGCATCTGATTAA
Protein
MATETLVPLESNPDVLNKFLQKLGVPNKWNIVDVMGLDPETLSWVPRPVLSVMLLFPISDAYENHKKTEENEILSKGQEVSGNIFYMKQNISNACGTIALVHSVANNTDIIELSDGHMQKFLNEAKGLDATARGKLLEKSEGIINAHKELAQEGQTNTPSAEDPVNHHFISFVQKDGALYELDGRKAFPVNHGPTSQETLLEDAAKICKEFMARDPNEVRFTVIALVASD
Summary
Description
Ubiquitin-protein hydrolase is involved both in the processing of ubiquitin precursors and of ubiquitinated proteins. This enzyme is a thiol protease that recognizes and hydrolyzes a peptide bond at the C-terminal glycine of ubiquitin.
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Similarity
Belongs to the peptidase C12 family.
Keywords
Complete proteome
Hydrolase
Protease
Reference proteome
Thiol protease
Ubl conjugation pathway
Feature
chain Ubiquitin carboxyl-terminal hydrolase
Uniprot
E5EVW1
I4DKT6
A0A2W1BL54
A0A2A4JUX1
A0A194RGS0
A0A2H1VVL3
+ More
S4NNT0 A0A0L7KLJ4 A0A212FJW0 R4V3Z4 A0A2J7PH84 A0A2J7PH73 A0A1B6L6K8 A0A1B6GNB3 E2B4G1 A0A1B6DXJ4 A0A158NQ48 A0A1Y1KU92 A0A2J7PH70 A0A2H1VCW9 A0A2A4K064 D6WF38 A0A1B0EWD8 H3ATA4 A0A2B4RRN0 A0A088ADV0 A0A1B6JRV9 A0A195BIZ1 A0A2A3E091 A0A1L8ED88 A0A151J061 A0A1B6LCB9 A0A212F4P9 A0A0V0G373 A0A1I8P8K9 A0A0C9QR99 A0A1B6FL35 J3JZJ6 A0A195FNM5 N6TNY7 A0A195CVR3 A0A1I8MWE8 W8BY65 A0A224XX86 A0A069DQC6 E2A1T2 A7S784 C3XWK4 A6YPQ5 U5EVC7 A0A0P4VYK6 R4G8N5 F4WMS1 E2J7E7 A0A1W4W4Z9 A0A3Q0J064 A0A3Q0J573 A0A067QU01 V9KNH4 A0A3M6TFU9 A0A0K8TT99 W5N056 Q5XHE2 A0A0A9YB44 K7J625 A0A0P4WK80 A0A194R1S8 A0A232F2K9 B4MVT2 A0A026WJW2 M9MRD0 P35122 R4WRB2 B3MJB3 D3TNH8 A0A1A9ZF47 C1K736 C4WTW9 A0A1A9UNR7 A0A1W5LJR1 A0A1B0C1N6 Q28GY5 A0A194Q7N1 A0A1W4V6Y9 E0V9V0 B4Q7N8 A0A1A9YIS3 A0A1B0FQ50 L5KKF6 A0A2Y9DBF8 A0A1A9W0Q2 A0A1U7STR8 A0A287CXP1 A0A2U3V1L6 A0A340WHQ4 A0A2Y9NCS4 A0A384AG15 G1LY51 G3SEB1 A0A2I3GN22
S4NNT0 A0A0L7KLJ4 A0A212FJW0 R4V3Z4 A0A2J7PH84 A0A2J7PH73 A0A1B6L6K8 A0A1B6GNB3 E2B4G1 A0A1B6DXJ4 A0A158NQ48 A0A1Y1KU92 A0A2J7PH70 A0A2H1VCW9 A0A2A4K064 D6WF38 A0A1B0EWD8 H3ATA4 A0A2B4RRN0 A0A088ADV0 A0A1B6JRV9 A0A195BIZ1 A0A2A3E091 A0A1L8ED88 A0A151J061 A0A1B6LCB9 A0A212F4P9 A0A0V0G373 A0A1I8P8K9 A0A0C9QR99 A0A1B6FL35 J3JZJ6 A0A195FNM5 N6TNY7 A0A195CVR3 A0A1I8MWE8 W8BY65 A0A224XX86 A0A069DQC6 E2A1T2 A7S784 C3XWK4 A6YPQ5 U5EVC7 A0A0P4VYK6 R4G8N5 F4WMS1 E2J7E7 A0A1W4W4Z9 A0A3Q0J064 A0A3Q0J573 A0A067QU01 V9KNH4 A0A3M6TFU9 A0A0K8TT99 W5N056 Q5XHE2 A0A0A9YB44 K7J625 A0A0P4WK80 A0A194R1S8 A0A232F2K9 B4MVT2 A0A026WJW2 M9MRD0 P35122 R4WRB2 B3MJB3 D3TNH8 A0A1A9ZF47 C1K736 C4WTW9 A0A1A9UNR7 A0A1W5LJR1 A0A1B0C1N6 Q28GY5 A0A194Q7N1 A0A1W4V6Y9 E0V9V0 B4Q7N8 A0A1A9YIS3 A0A1B0FQ50 L5KKF6 A0A2Y9DBF8 A0A1A9W0Q2 A0A1U7STR8 A0A287CXP1 A0A2U3V1L6 A0A340WHQ4 A0A2Y9NCS4 A0A384AG15 G1LY51 G3SEB1 A0A2I3GN22
EC Number
3.4.19.12
Pubmed
19121390
21457781
22651552
26354079
28756777
23622113
+ More
26227816 22118469 20798317 21347285 28004739 18362917 19820115 9215903 22516182 23537049 25315136 24495485 26334808 17615350 18563158 18207082 27129103 21719571 24845553 24402279 30382153 26369729 27762356 25401762 26823975 20075255 28648823 17994087 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1426603 7683284 10731138 23691247 20353571 20431018 20566863 22936249 23258410 20010809 22398555
26227816 22118469 20798317 21347285 28004739 18362917 19820115 9215903 22516182 23537049 25315136 24495485 26334808 17615350 18563158 18207082 27129103 21719571 24845553 24402279 30382153 26369729 27762356 25401762 26823975 20075255 28648823 17994087 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1426603 7683284 10731138 23691247 20353571 20431018 20566863 22936249 23258410 20010809 22398555
EMBL
BABH01014146
HM012805
ADQ89804.1
AK401904
KQ459589
BAM18526.1
+ More
KPI97514.1 KZ150098 PZC73586.1 NWSH01000598 PCG75424.1 KQ460207 KPJ16639.1 ODYU01004695 SOQ44853.1 GAIX01013816 JAA78744.1 JTDY01009043 KOB64167.1 AGBW02008206 OWR54028.1 KC740719 AGM32543.1 NEVH01025149 PNF15688.1 PNF15687.1 GEBQ01020650 JAT19327.1 GECZ01005831 JAS63938.1 GL445556 EFN89422.1 GEDC01006894 JAS30404.1 ADTU01022963 GEZM01075307 JAV64148.1 PNF15686.1 ODYU01001855 SOQ38636.1 NWSH01000333 PCG77318.1 KQ971327 EFA00904.1 AJWK01005593 AFYH01085089 AFYH01085090 AFYH01085091 AFYH01085092 AFYH01085093 AFYH01085094 AFYH01085095 AFYH01085096 AFYH01085097 LSMT01000359 PFX19459.1 GECU01005742 JAT01965.1 KQ976464 KYM84411.1 KZ288497 PBC25180.1 GFDG01002111 JAV16688.1 KQ980658 KYN14774.1 GEBQ01018650 JAT21327.1 AGBW02010334 OWR48703.1 GECL01003685 JAP02439.1 GBYB01003182 JAG72949.1 GECZ01018880 JAS50889.1 BT128678 KB632033 AEE63635.1 ERL88179.1 KQ981382 KYN42155.1 APGK01057490 KB741280 ENN70950.1 KQ977279 KYN04249.1 GAMC01008305 GAMC01008304 JAB98251.1 GFTR01003815 JAW12611.1 GBGD01002789 JAC86100.1 GL435798 EFN72604.1 DS469591 EDO40453.1 GG666471 EEN67738.1 EF639066 GEMB01003922 ABR27951.1 JAR99334.1 GANO01001091 JAB58780.1 GDKW01000892 JAI55703.1 ACPB03023923 GAHY01000662 JAA76848.1 GL888218 EGI64548.1 HP429365 ADN29865.1 KK853018 KDR12443.1 JW867635 AFP00153.1 RCHS01003667 RMX40276.1 GDAI01000463 JAI17140.1 AHAT01035001 BC084116 CM004468 AAH84116.1 OCT95509.1 GBHO01014210 GBRD01006179 GBRD01006178 GBRD01006177 GDHC01020228 JAG29394.1 JAG59642.1 JAP98400.1 GDRN01048972 JAI66623.1 KQ461073 KPJ09806.1 NNAY01001211 OXU24718.1 CH963857 EDW75802.1 KK107167 EZA56253.1 AE014134 ADV36932.1 S60346 X69678 X69679 AF145600 AK417212 BAN20427.1 CH902620 EDV31323.1 CCAG010011564 EZ422980 ADD19256.1 FJ800570 ACO36738.1 ABLF02030392 AK340802 BAH71339.1 KX091173 ANI85922.1 JXJN01024138 AAMC01006658 AAMC01006659 AAMC01006660 BC135703 CR761162 AAI35704.1 CAJ83746.1 KQ459324 KPJ01547.1 DS234996 EEB10156.1 CM000361 CM002910 EDX03418.1 KMY87606.1 CCAG010006700 KB030714 ELK11018.1 AGTP01036544 AGTP01036545 AGTP01036546 ACTA01180065 CABD030089848 CABD030089849 ADFV01174587
KPI97514.1 KZ150098 PZC73586.1 NWSH01000598 PCG75424.1 KQ460207 KPJ16639.1 ODYU01004695 SOQ44853.1 GAIX01013816 JAA78744.1 JTDY01009043 KOB64167.1 AGBW02008206 OWR54028.1 KC740719 AGM32543.1 NEVH01025149 PNF15688.1 PNF15687.1 GEBQ01020650 JAT19327.1 GECZ01005831 JAS63938.1 GL445556 EFN89422.1 GEDC01006894 JAS30404.1 ADTU01022963 GEZM01075307 JAV64148.1 PNF15686.1 ODYU01001855 SOQ38636.1 NWSH01000333 PCG77318.1 KQ971327 EFA00904.1 AJWK01005593 AFYH01085089 AFYH01085090 AFYH01085091 AFYH01085092 AFYH01085093 AFYH01085094 AFYH01085095 AFYH01085096 AFYH01085097 LSMT01000359 PFX19459.1 GECU01005742 JAT01965.1 KQ976464 KYM84411.1 KZ288497 PBC25180.1 GFDG01002111 JAV16688.1 KQ980658 KYN14774.1 GEBQ01018650 JAT21327.1 AGBW02010334 OWR48703.1 GECL01003685 JAP02439.1 GBYB01003182 JAG72949.1 GECZ01018880 JAS50889.1 BT128678 KB632033 AEE63635.1 ERL88179.1 KQ981382 KYN42155.1 APGK01057490 KB741280 ENN70950.1 KQ977279 KYN04249.1 GAMC01008305 GAMC01008304 JAB98251.1 GFTR01003815 JAW12611.1 GBGD01002789 JAC86100.1 GL435798 EFN72604.1 DS469591 EDO40453.1 GG666471 EEN67738.1 EF639066 GEMB01003922 ABR27951.1 JAR99334.1 GANO01001091 JAB58780.1 GDKW01000892 JAI55703.1 ACPB03023923 GAHY01000662 JAA76848.1 GL888218 EGI64548.1 HP429365 ADN29865.1 KK853018 KDR12443.1 JW867635 AFP00153.1 RCHS01003667 RMX40276.1 GDAI01000463 JAI17140.1 AHAT01035001 BC084116 CM004468 AAH84116.1 OCT95509.1 GBHO01014210 GBRD01006179 GBRD01006178 GBRD01006177 GDHC01020228 JAG29394.1 JAG59642.1 JAP98400.1 GDRN01048972 JAI66623.1 KQ461073 KPJ09806.1 NNAY01001211 OXU24718.1 CH963857 EDW75802.1 KK107167 EZA56253.1 AE014134 ADV36932.1 S60346 X69678 X69679 AF145600 AK417212 BAN20427.1 CH902620 EDV31323.1 CCAG010011564 EZ422980 ADD19256.1 FJ800570 ACO36738.1 ABLF02030392 AK340802 BAH71339.1 KX091173 ANI85922.1 JXJN01024138 AAMC01006658 AAMC01006659 AAMC01006660 BC135703 CR761162 AAI35704.1 CAJ83746.1 KQ459324 KPJ01547.1 DS234996 EEB10156.1 CM000361 CM002910 EDX03418.1 KMY87606.1 CCAG010006700 KB030714 ELK11018.1 AGTP01036544 AGTP01036545 AGTP01036546 ACTA01180065 CABD030089848 CABD030089849 ADFV01174587
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000037510
UP000007151
+ More
UP000235965 UP000008237 UP000005205 UP000007266 UP000092461 UP000008672 UP000225706 UP000005203 UP000078540 UP000242457 UP000078492 UP000095300 UP000030742 UP000078541 UP000019118 UP000078542 UP000095301 UP000000311 UP000001593 UP000001554 UP000015103 UP000007755 UP000192223 UP000079169 UP000027135 UP000275408 UP000018468 UP000186698 UP000002358 UP000215335 UP000007798 UP000053097 UP000000803 UP000007801 UP000092444 UP000092445 UP000007819 UP000078200 UP000092460 UP000008143 UP000192221 UP000009046 UP000000304 UP000092443 UP000010552 UP000248480 UP000091820 UP000189704 UP000005215 UP000245320 UP000265300 UP000248483 UP000261681 UP000008912 UP000001519 UP000001073
UP000235965 UP000008237 UP000005205 UP000007266 UP000092461 UP000008672 UP000225706 UP000005203 UP000078540 UP000242457 UP000078492 UP000095300 UP000030742 UP000078541 UP000019118 UP000078542 UP000095301 UP000000311 UP000001593 UP000001554 UP000015103 UP000007755 UP000192223 UP000079169 UP000027135 UP000275408 UP000018468 UP000186698 UP000002358 UP000215335 UP000007798 UP000053097 UP000000803 UP000007801 UP000092444 UP000092445 UP000007819 UP000078200 UP000092460 UP000008143 UP000192221 UP000009046 UP000000304 UP000092443 UP000010552 UP000248480 UP000091820 UP000189704 UP000005215 UP000245320 UP000265300 UP000248483 UP000261681 UP000008912 UP000001519 UP000001073
Pfam
PF01088 Peptidase_C12
Interpro
SUPFAM
SSF54001
SSF54001
Gene 3D
ProteinModelPortal
E5EVW1
I4DKT6
A0A2W1BL54
A0A2A4JUX1
A0A194RGS0
A0A2H1VVL3
+ More
S4NNT0 A0A0L7KLJ4 A0A212FJW0 R4V3Z4 A0A2J7PH84 A0A2J7PH73 A0A1B6L6K8 A0A1B6GNB3 E2B4G1 A0A1B6DXJ4 A0A158NQ48 A0A1Y1KU92 A0A2J7PH70 A0A2H1VCW9 A0A2A4K064 D6WF38 A0A1B0EWD8 H3ATA4 A0A2B4RRN0 A0A088ADV0 A0A1B6JRV9 A0A195BIZ1 A0A2A3E091 A0A1L8ED88 A0A151J061 A0A1B6LCB9 A0A212F4P9 A0A0V0G373 A0A1I8P8K9 A0A0C9QR99 A0A1B6FL35 J3JZJ6 A0A195FNM5 N6TNY7 A0A195CVR3 A0A1I8MWE8 W8BY65 A0A224XX86 A0A069DQC6 E2A1T2 A7S784 C3XWK4 A6YPQ5 U5EVC7 A0A0P4VYK6 R4G8N5 F4WMS1 E2J7E7 A0A1W4W4Z9 A0A3Q0J064 A0A3Q0J573 A0A067QU01 V9KNH4 A0A3M6TFU9 A0A0K8TT99 W5N056 Q5XHE2 A0A0A9YB44 K7J625 A0A0P4WK80 A0A194R1S8 A0A232F2K9 B4MVT2 A0A026WJW2 M9MRD0 P35122 R4WRB2 B3MJB3 D3TNH8 A0A1A9ZF47 C1K736 C4WTW9 A0A1A9UNR7 A0A1W5LJR1 A0A1B0C1N6 Q28GY5 A0A194Q7N1 A0A1W4V6Y9 E0V9V0 B4Q7N8 A0A1A9YIS3 A0A1B0FQ50 L5KKF6 A0A2Y9DBF8 A0A1A9W0Q2 A0A1U7STR8 A0A287CXP1 A0A2U3V1L6 A0A340WHQ4 A0A2Y9NCS4 A0A384AG15 G1LY51 G3SEB1 A0A2I3GN22
S4NNT0 A0A0L7KLJ4 A0A212FJW0 R4V3Z4 A0A2J7PH84 A0A2J7PH73 A0A1B6L6K8 A0A1B6GNB3 E2B4G1 A0A1B6DXJ4 A0A158NQ48 A0A1Y1KU92 A0A2J7PH70 A0A2H1VCW9 A0A2A4K064 D6WF38 A0A1B0EWD8 H3ATA4 A0A2B4RRN0 A0A088ADV0 A0A1B6JRV9 A0A195BIZ1 A0A2A3E091 A0A1L8ED88 A0A151J061 A0A1B6LCB9 A0A212F4P9 A0A0V0G373 A0A1I8P8K9 A0A0C9QR99 A0A1B6FL35 J3JZJ6 A0A195FNM5 N6TNY7 A0A195CVR3 A0A1I8MWE8 W8BY65 A0A224XX86 A0A069DQC6 E2A1T2 A7S784 C3XWK4 A6YPQ5 U5EVC7 A0A0P4VYK6 R4G8N5 F4WMS1 E2J7E7 A0A1W4W4Z9 A0A3Q0J064 A0A3Q0J573 A0A067QU01 V9KNH4 A0A3M6TFU9 A0A0K8TT99 W5N056 Q5XHE2 A0A0A9YB44 K7J625 A0A0P4WK80 A0A194R1S8 A0A232F2K9 B4MVT2 A0A026WJW2 M9MRD0 P35122 R4WRB2 B3MJB3 D3TNH8 A0A1A9ZF47 C1K736 C4WTW9 A0A1A9UNR7 A0A1W5LJR1 A0A1B0C1N6 Q28GY5 A0A194Q7N1 A0A1W4V6Y9 E0V9V0 B4Q7N8 A0A1A9YIS3 A0A1B0FQ50 L5KKF6 A0A2Y9DBF8 A0A1A9W0Q2 A0A1U7STR8 A0A287CXP1 A0A2U3V1L6 A0A340WHQ4 A0A2Y9NCS4 A0A384AG15 G1LY51 G3SEB1 A0A2I3GN22
PDB
6ISU
E-value=1.04563e-65,
Score=632
Ontologies
GO
PANTHER
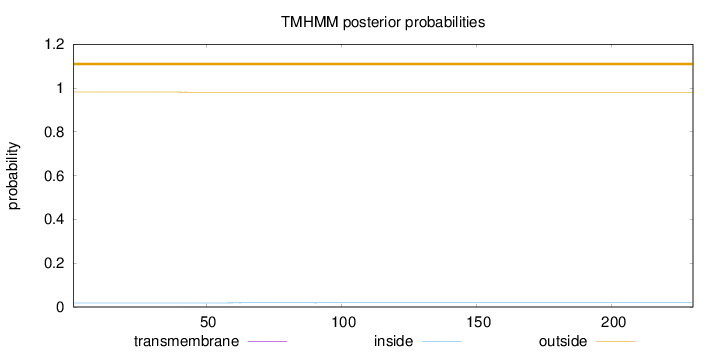
Topology
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01534
Exp number, first 60 AAs:
0.01189
Total prob of N-in:
0.01848
outside
1 - 230
Population Genetic Test Statistics
Pi
138.520051
Theta
137.720306
Tajima's D
0.018246
CLR
52.368979
CSRT
0.373131343432828
Interpretation
Uncertain
