Gene
KWMTBOMO02621
Pre Gene Modal
BGIBMGA002634
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.938
Sequence
CDS
ATGATACTTTGTTCAGCTGCGAGTCCAATTCCTGCATCTTACCTTGCCGATAGGATCGGAAGGAAGAAGACTCTTCTCCTGGCAGCAGTACCATACATCATCGGATGGATTCTAGTGATGCTTGCTGCAAACGTTCCTACAATTTACGCCTCTCGTCTCATCTCAGGGCTGGGTTACGGGATCGCCTACACTACAGCACCTATGTATCTCGGAGAGATAGCTTCAGATGAAGTTCGCGGGGCGATGGCAACCCTAATTACAGTAATGTCAAAGTTCGGCATCCTGTCGCAATATTGCATTGGGCCATACGTCTCAATGCTAGGTTTGGCAAGTTTTAACATAGCACTACCAATACTATTCGTTGTAACGTTCAGCGCAATGCCTGAGTCGCCGTATTACTACATCAAAGCAGGACTGAAAGACGAAGCAGAATTGTCGTTAAAGAAACTAAGAGGCAAACATTTTGTAAGAGACGAGCTAGAGAGTATGAACAGCTTAGTCACAGAAAACATGAAAAATAAAAGTCGGTGGAAAGATCTCATAACCGTTAGAGGAAACAAAAAAGGACTCATCATTCTTCTGGGAATTTATTTCACTCAACAGTTTTGCGGCAGTACTGCGATCATCAGCTATACACAGCAAATATTCGACGCCGCAGAAGGAGGATTAGGACCGGCTGAGTCTAGCATATTATTTGGTTCCGTGCAATTGTTAACTTCAGCAATCTCTTCACAATTAGTCGACAGATTAGGAAGGAAACCTCTACTATTAATATCTTCTTGTGGAGTGGCAGTGACCAACATCATAATCGGCGCTTACTTCTTCATAAAAGACTTGAACAGTGAATACGTCGTGAGCGTGAAATTCATTCCTTTAGTTGTAATACTAATATTTATATTTTCTTACACAATAGGCTTAGCCACTGTGCCTTTTGCTATTACTTCAGAGATGTTCCCCACTAATATTAAATCTAAAGCGACGTGTGTCATACAAATATTCGTGGCTTTGATAACATTTGCCGTCACGAAACTGTATCAAGTTGTGGATTCTAATTTAGGGACTTATGTGGTGTTCTGGGGTTTCGCGGTAATGTCAGTGGCCGGTGTGATATTTATTTTAATATTATTGCCAGAAACGAAAGGTCAATCGTTTGCGGCTATACAAGAGAAATTATATTCTAGTGAAAAAGTTGTTTACGAGAAAGACGAGGATCAGATGGCCAGTGTTAAAATAACATTGTGA
Protein
MILCSAASPIPASYLADRIGRKKTLLLAAVPYIIGWILVMLAANVPTIYASRLISGLGYGIAYTTAPMYLGEIASDEVRGAMATLITVMSKFGILSQYCIGPYVSMLGLASFNIALPILFVVTFSAMPESPYYYIKAGLKDEAELSLKKLRGKHFVRDELESMNSLVTENMKNKSRWKDLITVRGNKKGLIILLGIYFTQQFCGSTAIISYTQQIFDAAEGGLGPAESSILFGSVQLLTSAISSQLVDRLGRKPLLLISSCGVAVTNIIIGAYFFIKDLNSEYVVSVKFIPLVVILIFIFSYTIGLATVPFAITSEMFPTNIKSKATCVIQIFVALITFAVTKLYQVVDSNLGTYVVFWGFAVMSVAGVIFILILLPETKGQSFAAIQEKLYSSEKVVYEKDEDQMASVKITL
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9IZE9
A0A2A4JTT1
A0A2H1VF40
A0A194R1K9
A0A194PXP0
A0A212ERK6
+ More
A0A0L7LKK9 H9IZF0 A0A2A4JUE8 A0A212ERN0 A0A194PW18 A0A194R1W3 A0A2H1VY21 S4P7H5 A0A3G1T189 A0A0L7LJV7 A0A182PQF0 A0A182JUH6 A0A182X3Q9 Q7QJE9 A0A182V7I7 A0A182I3B0 A0A182XYW5 A0A182RSN7 A0A182S6W7 A0A182KRC0 A0A182TN07 A0A182J5I6 A0A1B0CUK2 A0A182NHV3 A0A182FF45 W5JRP7 A0A182WJ81 A0A182MUN7 A0A182QUI1 A0A084WNA8 Q16N91 B0X5T8 A0A1B0DJJ1 A0A1W7R5M1 A0A1B0DQK1 A0A067R7D8 A0A2J7RK51 A0A182G3D2 Q7QJF0 A0A182KRB6 U5ESS0 A0A182VHI4 A0A182X3Q8 E9IVK0 A0A182IRD4 T1DE67 A0A182I3B1 A0A2J7PYS8 A0A182K3E7 A0A182J8U1 E2ALN5 A0A0A8J7X6 A0A182PUD6 A0A084WNA9 A0A182JUH7 A0A182MWN6 A0A182YRA3 A0A1Q3FQH2 A0A1Q3FQA6 B0X5T9 A0A182RLR4 A0A182SQT4 A0A0K8W987 A0A1Q3FQG8 A0A1Q3FQD3 E2ADH1 A0A0K8WL46 Q16SU5 A0A182S4P1 A0A0K8U506 A0A182NLK4 W8B3Q4 A0A182GL69 A0A0M8ZMY1 A0A182LZZ1 A0A026WP92 A0A182FF46 B0WZW7 W8BDP5 A0A1Y1MRV1 A0A3L8DJN2 A0A182GVZ1 A0A182GCW7 A0A182QT48 A0A084VDW1 E9IQR3 B0WDB0 W5J543 A0A1S4FXY4 Q16KS3 T1DG93 A0A182QHE6 A0A3L8DXN6
A0A0L7LKK9 H9IZF0 A0A2A4JUE8 A0A212ERN0 A0A194PW18 A0A194R1W3 A0A2H1VY21 S4P7H5 A0A3G1T189 A0A0L7LJV7 A0A182PQF0 A0A182JUH6 A0A182X3Q9 Q7QJE9 A0A182V7I7 A0A182I3B0 A0A182XYW5 A0A182RSN7 A0A182S6W7 A0A182KRC0 A0A182TN07 A0A182J5I6 A0A1B0CUK2 A0A182NHV3 A0A182FF45 W5JRP7 A0A182WJ81 A0A182MUN7 A0A182QUI1 A0A084WNA8 Q16N91 B0X5T8 A0A1B0DJJ1 A0A1W7R5M1 A0A1B0DQK1 A0A067R7D8 A0A2J7RK51 A0A182G3D2 Q7QJF0 A0A182KRB6 U5ESS0 A0A182VHI4 A0A182X3Q8 E9IVK0 A0A182IRD4 T1DE67 A0A182I3B1 A0A2J7PYS8 A0A182K3E7 A0A182J8U1 E2ALN5 A0A0A8J7X6 A0A182PUD6 A0A084WNA9 A0A182JUH7 A0A182MWN6 A0A182YRA3 A0A1Q3FQH2 A0A1Q3FQA6 B0X5T9 A0A182RLR4 A0A182SQT4 A0A0K8W987 A0A1Q3FQG8 A0A1Q3FQD3 E2ADH1 A0A0K8WL46 Q16SU5 A0A182S4P1 A0A0K8U506 A0A182NLK4 W8B3Q4 A0A182GL69 A0A0M8ZMY1 A0A182LZZ1 A0A026WP92 A0A182FF46 B0WZW7 W8BDP5 A0A1Y1MRV1 A0A3L8DJN2 A0A182GVZ1 A0A182GCW7 A0A182QT48 A0A084VDW1 E9IQR3 B0WDB0 W5J543 A0A1S4FXY4 Q16KS3 T1DG93 A0A182QHE6 A0A3L8DXN6
Pubmed
EMBL
BABH01014162
NWSH01000598
PCG75421.1
ODYU01001965
SOQ38884.1
KQ460870
+ More
KPJ11683.1 KQ459589 KPI97524.1 AGBW02012985 OWR44136.1 JTDY01000837 KOB75721.1 BABH01014157 PCG75429.1 OWR44135.1 KPI97522.1 KPJ11682.1 ODYU01004836 SOQ45124.1 GAIX01004609 JAA87951.1 MG846896 AXY94748.1 KOB75722.1 AAAB01008807 EAA04375.4 APCN01000204 AJWK01029294 ADMH02000549 ETN65958.1 AXCM01011849 AXCN02000379 ATLV01024580 KE525352 KFB51702.1 CH477833 EAT35815.1 DS232396 EDS41069.1 AJVK01035036 AJVK01035037 GEHC01001176 JAV46469.1 AJVK01008622 AJVK01008623 AJVK01008624 KK852657 KDR19246.1 NEVH01002981 PNF41213.1 JXUM01040843 KQ561261 KXJ79172.1 EAA04637.4 GANO01003127 JAB56744.1 GL766258 EFZ15414.1 GALA01001167 JAA93685.1 NEVH01020342 PNF21493.1 GL440609 EFN65644.1 AB901052 BAQ02352.1 KFB51703.1 AXCM01016880 GFDL01005283 JAV29762.1 GFDL01005255 JAV29790.1 EDS41070.1 GDHF01004672 JAI47642.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GL438750 EFN68533.1 GDHF01000517 JAI51797.1 CH477665 EAT37543.1 GDHF01030597 JAI21717.1 GAMC01018654 JAB87901.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 KQ436022 KOX67560.1 AXCM01019063 KK107139 EZA57783.1 DS232219 EDS37792.1 GAMC01018656 JAB87899.1 GEZM01023497 JAV88361.1 QOIP01000007 RLU20546.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 JXUM01054849 KQ561839 KXJ77384.1 AXCN02000090 ATLV01011809 KE524723 KFB36155.1 GL764900 EFZ17069.1 DS231895 EDS44344.1 ADMH02002130 ETN58528.1 CH477941 EAT34897.1 GAMD01002846 JAA98744.1 QOIP01000003 RLU25240.1
KPJ11683.1 KQ459589 KPI97524.1 AGBW02012985 OWR44136.1 JTDY01000837 KOB75721.1 BABH01014157 PCG75429.1 OWR44135.1 KPI97522.1 KPJ11682.1 ODYU01004836 SOQ45124.1 GAIX01004609 JAA87951.1 MG846896 AXY94748.1 KOB75722.1 AAAB01008807 EAA04375.4 APCN01000204 AJWK01029294 ADMH02000549 ETN65958.1 AXCM01011849 AXCN02000379 ATLV01024580 KE525352 KFB51702.1 CH477833 EAT35815.1 DS232396 EDS41069.1 AJVK01035036 AJVK01035037 GEHC01001176 JAV46469.1 AJVK01008622 AJVK01008623 AJVK01008624 KK852657 KDR19246.1 NEVH01002981 PNF41213.1 JXUM01040843 KQ561261 KXJ79172.1 EAA04637.4 GANO01003127 JAB56744.1 GL766258 EFZ15414.1 GALA01001167 JAA93685.1 NEVH01020342 PNF21493.1 GL440609 EFN65644.1 AB901052 BAQ02352.1 KFB51703.1 AXCM01016880 GFDL01005283 JAV29762.1 GFDL01005255 JAV29790.1 EDS41070.1 GDHF01004672 JAI47642.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GL438750 EFN68533.1 GDHF01000517 JAI51797.1 CH477665 EAT37543.1 GDHF01030597 JAI21717.1 GAMC01018654 JAB87901.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 KQ436022 KOX67560.1 AXCM01019063 KK107139 EZA57783.1 DS232219 EDS37792.1 GAMC01018656 JAB87899.1 GEZM01023497 JAV88361.1 QOIP01000007 RLU20546.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 JXUM01054849 KQ561839 KXJ77384.1 AXCN02000090 ATLV01011809 KE524723 KFB36155.1 GL764900 EFZ17069.1 DS231895 EDS44344.1 ADMH02002130 ETN58528.1 CH477941 EAT34897.1 GAMD01002846 JAA98744.1 QOIP01000003 RLU25240.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000075885 UP000075881 UP000076407 UP000007062 UP000075903 UP000075840 UP000076408 UP000075900 UP000075901 UP000075882 UP000075902 UP000075880 UP000092461 UP000075884 UP000069272 UP000000673 UP000075920 UP000075883 UP000075886 UP000030765 UP000008820 UP000002320 UP000092462 UP000027135 UP000235965 UP000069940 UP000249989 UP000000311 UP000053105 UP000053097 UP000279307
UP000075885 UP000075881 UP000076407 UP000007062 UP000075903 UP000075840 UP000076408 UP000075900 UP000075901 UP000075882 UP000075902 UP000075880 UP000092461 UP000075884 UP000069272 UP000000673 UP000075920 UP000075883 UP000075886 UP000030765 UP000008820 UP000002320 UP000092462 UP000027135 UP000235965 UP000069940 UP000249989 UP000000311 UP000053105 UP000053097 UP000279307
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IZE9
A0A2A4JTT1
A0A2H1VF40
A0A194R1K9
A0A194PXP0
A0A212ERK6
+ More
A0A0L7LKK9 H9IZF0 A0A2A4JUE8 A0A212ERN0 A0A194PW18 A0A194R1W3 A0A2H1VY21 S4P7H5 A0A3G1T189 A0A0L7LJV7 A0A182PQF0 A0A182JUH6 A0A182X3Q9 Q7QJE9 A0A182V7I7 A0A182I3B0 A0A182XYW5 A0A182RSN7 A0A182S6W7 A0A182KRC0 A0A182TN07 A0A182J5I6 A0A1B0CUK2 A0A182NHV3 A0A182FF45 W5JRP7 A0A182WJ81 A0A182MUN7 A0A182QUI1 A0A084WNA8 Q16N91 B0X5T8 A0A1B0DJJ1 A0A1W7R5M1 A0A1B0DQK1 A0A067R7D8 A0A2J7RK51 A0A182G3D2 Q7QJF0 A0A182KRB6 U5ESS0 A0A182VHI4 A0A182X3Q8 E9IVK0 A0A182IRD4 T1DE67 A0A182I3B1 A0A2J7PYS8 A0A182K3E7 A0A182J8U1 E2ALN5 A0A0A8J7X6 A0A182PUD6 A0A084WNA9 A0A182JUH7 A0A182MWN6 A0A182YRA3 A0A1Q3FQH2 A0A1Q3FQA6 B0X5T9 A0A182RLR4 A0A182SQT4 A0A0K8W987 A0A1Q3FQG8 A0A1Q3FQD3 E2ADH1 A0A0K8WL46 Q16SU5 A0A182S4P1 A0A0K8U506 A0A182NLK4 W8B3Q4 A0A182GL69 A0A0M8ZMY1 A0A182LZZ1 A0A026WP92 A0A182FF46 B0WZW7 W8BDP5 A0A1Y1MRV1 A0A3L8DJN2 A0A182GVZ1 A0A182GCW7 A0A182QT48 A0A084VDW1 E9IQR3 B0WDB0 W5J543 A0A1S4FXY4 Q16KS3 T1DG93 A0A182QHE6 A0A3L8DXN6
A0A0L7LKK9 H9IZF0 A0A2A4JUE8 A0A212ERN0 A0A194PW18 A0A194R1W3 A0A2H1VY21 S4P7H5 A0A3G1T189 A0A0L7LJV7 A0A182PQF0 A0A182JUH6 A0A182X3Q9 Q7QJE9 A0A182V7I7 A0A182I3B0 A0A182XYW5 A0A182RSN7 A0A182S6W7 A0A182KRC0 A0A182TN07 A0A182J5I6 A0A1B0CUK2 A0A182NHV3 A0A182FF45 W5JRP7 A0A182WJ81 A0A182MUN7 A0A182QUI1 A0A084WNA8 Q16N91 B0X5T8 A0A1B0DJJ1 A0A1W7R5M1 A0A1B0DQK1 A0A067R7D8 A0A2J7RK51 A0A182G3D2 Q7QJF0 A0A182KRB6 U5ESS0 A0A182VHI4 A0A182X3Q8 E9IVK0 A0A182IRD4 T1DE67 A0A182I3B1 A0A2J7PYS8 A0A182K3E7 A0A182J8U1 E2ALN5 A0A0A8J7X6 A0A182PUD6 A0A084WNA9 A0A182JUH7 A0A182MWN6 A0A182YRA3 A0A1Q3FQH2 A0A1Q3FQA6 B0X5T9 A0A182RLR4 A0A182SQT4 A0A0K8W987 A0A1Q3FQG8 A0A1Q3FQD3 E2ADH1 A0A0K8WL46 Q16SU5 A0A182S4P1 A0A0K8U506 A0A182NLK4 W8B3Q4 A0A182GL69 A0A0M8ZMY1 A0A182LZZ1 A0A026WP92 A0A182FF46 B0WZW7 W8BDP5 A0A1Y1MRV1 A0A3L8DJN2 A0A182GVZ1 A0A182GCW7 A0A182QT48 A0A084VDW1 E9IQR3 B0WDB0 W5J543 A0A1S4FXY4 Q16KS3 T1DG93 A0A182QHE6 A0A3L8DXN6
PDB
4LDS
E-value=3.35936e-31,
Score=337
Ontologies
GO
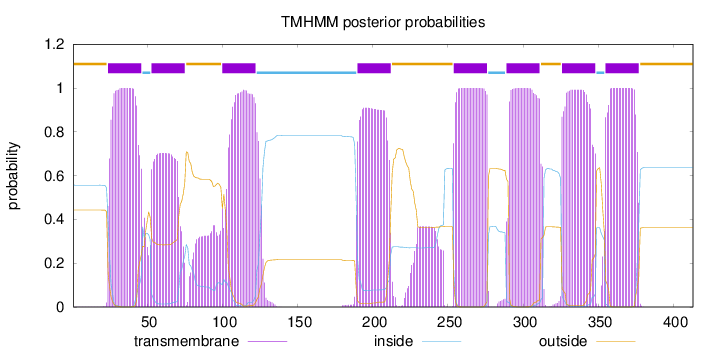
Topology
Length:
413
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
183.91328
Exp number, first 60 AAs:
28.16242
Total prob of N-in:
0.55688
POSSIBLE N-term signal
sequence
outside
1 - 23
TMhelix
24 - 46
inside
47 - 52
TMhelix
53 - 75
outside
76 - 99
TMhelix
100 - 122
inside
123 - 189
TMhelix
190 - 212
outside
213 - 253
TMhelix
254 - 276
inside
277 - 288
TMhelix
289 - 311
outside
312 - 325
TMhelix
326 - 348
inside
349 - 354
TMhelix
355 - 377
outside
378 - 413
Population Genetic Test Statistics
Pi
248.712934
Theta
189.561799
Tajima's D
1.131079
CLR
0.401796
CSRT
0.695815209239538
Interpretation
Uncertain
