Gene
KWMTBOMO02617 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002661
Annotation
PREDICTED:_lipophorin_receptor_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.833
Sequence
CDS
ATGTATCTACGGGTGTGTATCCTGATTTTTACATTCATCGTTATCACGGAAGCGACGTTTTACGATGACGATTACGACAGCTACACCAAGGCTTACGACTACAAAAGTGCAGAGAGCCGCACTTGCACGTCCTCCGAGTTCCGTTGCAAAACTGGAAGATGCATACCATTATCGTGGAAATGTGACAACGAAAAAGATTGTTCGGATAGTTCTGACGAGGACCCTACCGTCTGCAAAGTGGAAGCTTGCGGCCCGGAGGAGTTCACGTGCAGGGGAAAGCCTGGTGAATGTGTCCCACTCACTTGGATGTGCGACGACAATCCCGATTGCTCCGATGGATCAGATGAAAAAGCATGCAACGAGACTTGTCGTTCCGATGAGTTCACCTGTGGGAATGGGAAGTGCATTCAAATGCGATGGGTGTGCGATGGTGACGACGATTGCGGCGATGACAGCGACGAAGTGAAATGTCCAACCCCAACTTGCCAGCCGCAGTCGCACTTCTCATGTACAGATGGTCAGTGTATATCAGCGTGGTGGCACTGTGATGGTGACGTCGACTGTGCCGATGGAAGCGATGAAGTGGGATGCGTCTCCACACCTAAAGTCAGATCTCCGTGTATTTCTACCGAGTTCGAATGCAACGATCGCATAACCTGTGTGCACCGGGCCTGGGTCTGTGACGGTGACCACGATTGTCCCGATGGTGGTGACGAAGCTCTTGAACTATGTCGGGGCAATGTCACTTGCAGACTAGATCAATTCCAATGCAAGGACCATAGCTGTATTCCGGGAGCCCTCTATTGTAACGGGGTTAAGGACTGCCCTGATGGCAGCGATGAGTACAACTGCACCCGACAAAAAACTATTTGCGACAAAAGAACAGAATTTGACTGCGGAGGCGGTATGTGCATCCCATTGTCCAAAGTTTGTGATAAACATGTGGACTGCCCCAACTTTGAGGACGAGCCAAGAGACAGATGCGGTGAGAACGAATGTGAGAAGAACAACGGTGGATGCTCTCAACGTTGTGTGGATACACCAATTGGATACTATTGTGATTGTGAAAAGGGCTACAAGCTTATCGACAACAGGACTTGTGAAGATATAGACGAATGCGCGGACCCTGGATCTTGCTCACAAATATGCATTAACGAAAAAGGCACCTTCAAATGCGAATGTCATACCGGCTACGCCAGAGACCCTCGTGACAGAACCAGGTGCAAAGCAACGGAGGGGCACCCATCATTACTGTTCGCAAGAAAACACGACATTCGTAAGATAAGCCTGGACCATCATGAAATGGTGGCGATTGTGAACGACACAAAATCAGCAACGGCACTCGACTATGTCTTCAGAACCGGAATGATCTTTTGGAGCGATGTCAATGACGAGAAGATTTACAAAGCACCAATTGATGAAGGCACTCAACGTACAGTCGTCATCGGTGAACAGCTGATTACATCCGACGGCCTCGCAGTCGACTGGGTCTATAATCATCTGTATTGGACCGACACCGGGCACAACCACATCGAGCTCTCCGATTTACAAGGGAACATGAGAAAAATACTGATCAGAGATCGCCTGGAAGAACCACGTGCAATTGCACTGAATCCTTTAGAAGGTTGGATGTTCTGGACCGACTGGGGTCAAGTGCCGAAGATCGAACGTGCTGGCATGGACGGTTCACATCGGAGCACCATTGTGTCTTACGACGTAAAATGGCCCAACGGTCTGACTCTGGACCTTGTACGCCAACGAGTTTACTGGGTGGATGCGAAAATGAATACAATCTCATCGTGCAACTACGACGGGTCCGGGCGCCGGCTCATTCTCCATTCGACCGAAGTGTTGAGACACCCGTTCTCGATAACCACATTCGAAGACTGGGTCTACTGGACTGATTGGGACAAGACAGCCGTGTACCGAGCGAATAAATTCAACGGTAAAGACGTGGAAGCAATCACATCTACACACACGGTGAGTATTTAA
Protein
MYLRVCILIFTFIVITEATFYDDDYDSYTKAYDYKSAESRTCTSSEFRCKTGRCIPLSWKCDNEKDCSDSSDEDPTVCKVEACGPEEFTCRGKPGECVPLTWMCDDNPDCSDGSDEKACNETCRSDEFTCGNGKCIQMRWVCDGDDDCGDDSDEVKCPTPTCQPQSHFSCTDGQCISAWWHCDGDVDCADGSDEVGCVSTPKVRSPCISTEFECNDRITCVHRAWVCDGDHDCPDGGDEALELCRGNVTCRLDQFQCKDHSCIPGALYCNGVKDCPDGSDEYNCTRQKTICDKRTEFDCGGGMCIPLSKVCDKHVDCPNFEDEPRDRCGENECEKNNGGCSQRCVDTPIGYYCDCEKGYKLIDNRTCEDIDECADPGSCSQICINEKGTFKCECHTGYARDPRDRTRCKATEGHPSLLFARKHDIRKISLDHHEMVAIVNDTKSATALDYVFRTGMIFWSDVNDEKIYKAPIDEGTQRTVVIGEQLITSDGLAVDWVYNHLYWTDTGHNHIELSDLQGNMRKILIRDRLEEPRAIALNPLEGWMFWTDWGQVPKIERAGMDGSHRSTIVSYDVKWPNGLTLDLVRQRVYWVDAKMNTISSCNYDGSGRRLILHSTEVLRHPFSITTFEDWVYWTDWDKTAVYRANKFNGKDVEAITSTHTVSI
Summary
Uniprot
Q2PGW2
Q2PGW1
Q2PGV9
A4PBE1
H9IZH6
A0A1B0Z2Q9
+ More
A0A1B0Z2Q6 Q2PGW0 Q1HI61 A0A194Q293 A0A1Z2RRF4 A0A1E1WNF2 G4V3R2 A0A194R2D8 A0A173DR70 A3KL83 A3KL84 Q4R1B4 O46131 A0A2J7R3N9 A0A2J7R3M7 A0A1Y1LSU7 A0A1Y1LRX7 A0A139WA84 E0VQJ0 A0A1L8E3G6 A0A1L8E429 A0A1J1III2 A0A023EW69 A0A0T6B2I0 D5KXW5 A0A1Q3G426 A0A1Q3G483 Q963T3 Q7YW57 A0A2A4J5C8 A0A1Q3G4D8 A0A1Q3G478 A0A2M4CVC1 A0A1Q3G465 A0A0V0G997 A0A2M4BD43 A0A2M3ZEA8 A0A2M4BDU5 A0A2M3Z2J9 A0A2M3Z2K2 A0A1Q3G435 A0A1Q3G473 Q7QK77 A0A1Q3G4A2 F5HIV0 T1PEP9 A0A1I8M8M8 T1PLU1 A0A1J1IF34 T1PP38 A0A2J7R3N4 T1PLT6 W8B062 U5ER34 A0A1L8E3H2 A0A1L8E3V1 W8BF58 A0A2M4CVB8 B3LYM0 A0A034VFQ6 A0A2M4DLP2 A0A1I8M8M3 A0A0P8YMD8 A0A1I8M8N1 A0A0P8XZK3 A0A1I8M8M5 A0A0P9AR17 A0A0P8YF07 A0A0K8UZB7 B4K4U9 A0A034VE53 A0A0Q9XBK3 A0A0Q9X1I1 A0A0R3NJ46 A0A0R3NIZ5 A0A1I8M8N3 I5APG6 A0A224X707 A0A0Q9X1I8 A0A1W4VLK6 A0A1W4VK86 A0A1W4V8H5 A0A0M5J046 A0A1W4VLM1 A0A1W4V833 A0A0R3NP71 B4K4V0 A0A0R3NJ76 I5APG7 A0A0Q9X257 A0A0Q9XE49 B3P6F8 A0A1I8M8N4
A0A1B0Z2Q6 Q2PGW0 Q1HI61 A0A194Q293 A0A1Z2RRF4 A0A1E1WNF2 G4V3R2 A0A194R2D8 A0A173DR70 A3KL83 A3KL84 Q4R1B4 O46131 A0A2J7R3N9 A0A2J7R3M7 A0A1Y1LSU7 A0A1Y1LRX7 A0A139WA84 E0VQJ0 A0A1L8E3G6 A0A1L8E429 A0A1J1III2 A0A023EW69 A0A0T6B2I0 D5KXW5 A0A1Q3G426 A0A1Q3G483 Q963T3 Q7YW57 A0A2A4J5C8 A0A1Q3G4D8 A0A1Q3G478 A0A2M4CVC1 A0A1Q3G465 A0A0V0G997 A0A2M4BD43 A0A2M3ZEA8 A0A2M4BDU5 A0A2M3Z2J9 A0A2M3Z2K2 A0A1Q3G435 A0A1Q3G473 Q7QK77 A0A1Q3G4A2 F5HIV0 T1PEP9 A0A1I8M8M8 T1PLU1 A0A1J1IF34 T1PP38 A0A2J7R3N4 T1PLT6 W8B062 U5ER34 A0A1L8E3H2 A0A1L8E3V1 W8BF58 A0A2M4CVB8 B3LYM0 A0A034VFQ6 A0A2M4DLP2 A0A1I8M8M3 A0A0P8YMD8 A0A1I8M8N1 A0A0P8XZK3 A0A1I8M8M5 A0A0P9AR17 A0A0P8YF07 A0A0K8UZB7 B4K4U9 A0A034VE53 A0A0Q9XBK3 A0A0Q9X1I1 A0A0R3NJ46 A0A0R3NIZ5 A0A1I8M8N3 I5APG6 A0A224X707 A0A0Q9X1I8 A0A1W4VLK6 A0A1W4VK86 A0A1W4V8H5 A0A0M5J046 A0A1W4VLM1 A0A1W4V833 A0A0R3NP71 B4K4V0 A0A0R3NJ76 I5APG7 A0A0Q9X257 A0A0Q9XE49 B3P6F8 A0A1I8M8N4
Pubmed
EMBL
AB201471
BAE71406.1
AB201472
BAE71407.1
AB201474
BAE71409.1
+ More
AB211594 BAF51965.1 BABH01014177 BABH01014178 KU936050 ANO58556.1 KU936051 ANO58557.1 AB201473 BAE71408.1 DQ482581 ABF20542.1 KQ459589 KPI97525.1 KY924787 ASA46441.1 GDQN01002688 JAT88366.1 GU433377 ADN04911.1 KQ460870 KPJ11684.1 KU355886 ANG56609.1 AM403063 CAL47125.1 AM403064 CAL47126.1 AB218869 BAE00010.1 AJ000010 CAA03855.1 NEVH01007818 PNF35442.1 PNF35444.1 GEZM01051892 JAV74646.1 GEZM01051893 JAV74645.1 KQ971466 KYB24825.1 DS235430 EEB15646.1 GFDF01000801 JAV13283.1 GFDF01000802 JAV13282.1 CVRI01000048 CRK98878.1 GAPW01000158 JAC13440.1 LJIG01016084 KRT81663.1 GU723298 ADE34167.2 GFDL01000494 JAV34551.1 GFDL01000434 JAV34611.1 AF355595 AAK72954.1 AY348869 JN411069 AAQ16410.1 AEY84776.1 NWSH01003102 PCG66946.1 GFDL01000419 JAV34626.1 GFDL01000438 JAV34607.1 GGFL01005105 MBW69283.1 GFDL01000452 JAV34593.1 GECL01002279 JAP03845.1 GGFJ01001823 MBW50964.1 GGFM01006019 MBW26770.1 GGFJ01002089 MBW51230.1 GGFM01001993 MBW22744.1 GGFM01001989 MBW22740.1 GFDL01000470 JAV34575.1 GFDL01000445 JAV34600.1 AAAB01008799 EAA03795.5 GFDL01000414 JAV34631.1 EGK96211.1 KA647257 KA649760 AFP61886.1 KA649757 KA649761 AFP64390.1 CRK98877.1 KA648951 KA649758 AFP64387.1 PNF35443.1 KA649756 KA649759 AFP64385.1 GAMC01014643 JAB91912.1 GANO01002986 JAB56885.1 GFDF01000794 JAV13290.1 GFDF01000795 JAV13289.1 GAMC01014644 JAB91911.1 GGFL01005106 MBW69284.1 CH902617 EDV42935.2 GAKP01018559 GAKP01018557 JAC40393.1 GGFL01014281 MBW78459.1 KPU79987.1 KPU79990.1 KPU79992.1 KPU79984.1 KPU79991.1 GDHF01020297 JAI32017.1 CH933806 EDW16102.1 GAKP01018560 JAC40392.1 KRG01934.1 KRG01935.1 CM000070 KRT00951.1 KRT00952.1 EIM52851.2 GFTR01008230 JAW08196.1 KRG01932.1 CP012526 ALC46956.1 KRT00949.1 EDW16103.2 KRT00948.1 EIM52852.2 KRG01941.1 KRG01939.1 CH954182 EDV53628.1
AB211594 BAF51965.1 BABH01014177 BABH01014178 KU936050 ANO58556.1 KU936051 ANO58557.1 AB201473 BAE71408.1 DQ482581 ABF20542.1 KQ459589 KPI97525.1 KY924787 ASA46441.1 GDQN01002688 JAT88366.1 GU433377 ADN04911.1 KQ460870 KPJ11684.1 KU355886 ANG56609.1 AM403063 CAL47125.1 AM403064 CAL47126.1 AB218869 BAE00010.1 AJ000010 CAA03855.1 NEVH01007818 PNF35442.1 PNF35444.1 GEZM01051892 JAV74646.1 GEZM01051893 JAV74645.1 KQ971466 KYB24825.1 DS235430 EEB15646.1 GFDF01000801 JAV13283.1 GFDF01000802 JAV13282.1 CVRI01000048 CRK98878.1 GAPW01000158 JAC13440.1 LJIG01016084 KRT81663.1 GU723298 ADE34167.2 GFDL01000494 JAV34551.1 GFDL01000434 JAV34611.1 AF355595 AAK72954.1 AY348869 JN411069 AAQ16410.1 AEY84776.1 NWSH01003102 PCG66946.1 GFDL01000419 JAV34626.1 GFDL01000438 JAV34607.1 GGFL01005105 MBW69283.1 GFDL01000452 JAV34593.1 GECL01002279 JAP03845.1 GGFJ01001823 MBW50964.1 GGFM01006019 MBW26770.1 GGFJ01002089 MBW51230.1 GGFM01001993 MBW22744.1 GGFM01001989 MBW22740.1 GFDL01000470 JAV34575.1 GFDL01000445 JAV34600.1 AAAB01008799 EAA03795.5 GFDL01000414 JAV34631.1 EGK96211.1 KA647257 KA649760 AFP61886.1 KA649757 KA649761 AFP64390.1 CRK98877.1 KA648951 KA649758 AFP64387.1 PNF35443.1 KA649756 KA649759 AFP64385.1 GAMC01014643 JAB91912.1 GANO01002986 JAB56885.1 GFDF01000794 JAV13290.1 GFDF01000795 JAV13289.1 GAMC01014644 JAB91911.1 GGFL01005106 MBW69284.1 CH902617 EDV42935.2 GAKP01018559 GAKP01018557 JAC40393.1 GGFL01014281 MBW78459.1 KPU79987.1 KPU79990.1 KPU79992.1 KPU79984.1 KPU79991.1 GDHF01020297 JAI32017.1 CH933806 EDW16102.1 GAKP01018560 JAC40392.1 KRG01934.1 KRG01935.1 CM000070 KRT00951.1 KRT00952.1 EIM52851.2 GFTR01008230 JAW08196.1 KRG01932.1 CP012526 ALC46956.1 KRT00949.1 EDW16103.2 KRT00948.1 EIM52852.2 KRG01941.1 KRG01939.1 CH954182 EDV53628.1
Proteomes
Interpro
IPR002172
LDrepeatLR_classA_rpt
+ More
IPR036055 LDL_receptor-like_sf
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR023415 LDLR_class-A_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR026823 cEGF
IPR036055 LDL_receptor-like_sf
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR023415 LDLR_class-A_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR026823 cEGF
Gene 3D
CDD
ProteinModelPortal
Q2PGW2
Q2PGW1
Q2PGV9
A4PBE1
H9IZH6
A0A1B0Z2Q9
+ More
A0A1B0Z2Q6 Q2PGW0 Q1HI61 A0A194Q293 A0A1Z2RRF4 A0A1E1WNF2 G4V3R2 A0A194R2D8 A0A173DR70 A3KL83 A3KL84 Q4R1B4 O46131 A0A2J7R3N9 A0A2J7R3M7 A0A1Y1LSU7 A0A1Y1LRX7 A0A139WA84 E0VQJ0 A0A1L8E3G6 A0A1L8E429 A0A1J1III2 A0A023EW69 A0A0T6B2I0 D5KXW5 A0A1Q3G426 A0A1Q3G483 Q963T3 Q7YW57 A0A2A4J5C8 A0A1Q3G4D8 A0A1Q3G478 A0A2M4CVC1 A0A1Q3G465 A0A0V0G997 A0A2M4BD43 A0A2M3ZEA8 A0A2M4BDU5 A0A2M3Z2J9 A0A2M3Z2K2 A0A1Q3G435 A0A1Q3G473 Q7QK77 A0A1Q3G4A2 F5HIV0 T1PEP9 A0A1I8M8M8 T1PLU1 A0A1J1IF34 T1PP38 A0A2J7R3N4 T1PLT6 W8B062 U5ER34 A0A1L8E3H2 A0A1L8E3V1 W8BF58 A0A2M4CVB8 B3LYM0 A0A034VFQ6 A0A2M4DLP2 A0A1I8M8M3 A0A0P8YMD8 A0A1I8M8N1 A0A0P8XZK3 A0A1I8M8M5 A0A0P9AR17 A0A0P8YF07 A0A0K8UZB7 B4K4U9 A0A034VE53 A0A0Q9XBK3 A0A0Q9X1I1 A0A0R3NJ46 A0A0R3NIZ5 A0A1I8M8N3 I5APG6 A0A224X707 A0A0Q9X1I8 A0A1W4VLK6 A0A1W4VK86 A0A1W4V8H5 A0A0M5J046 A0A1W4VLM1 A0A1W4V833 A0A0R3NP71 B4K4V0 A0A0R3NJ76 I5APG7 A0A0Q9X257 A0A0Q9XE49 B3P6F8 A0A1I8M8N4
A0A1B0Z2Q6 Q2PGW0 Q1HI61 A0A194Q293 A0A1Z2RRF4 A0A1E1WNF2 G4V3R2 A0A194R2D8 A0A173DR70 A3KL83 A3KL84 Q4R1B4 O46131 A0A2J7R3N9 A0A2J7R3M7 A0A1Y1LSU7 A0A1Y1LRX7 A0A139WA84 E0VQJ0 A0A1L8E3G6 A0A1L8E429 A0A1J1III2 A0A023EW69 A0A0T6B2I0 D5KXW5 A0A1Q3G426 A0A1Q3G483 Q963T3 Q7YW57 A0A2A4J5C8 A0A1Q3G4D8 A0A1Q3G478 A0A2M4CVC1 A0A1Q3G465 A0A0V0G997 A0A2M4BD43 A0A2M3ZEA8 A0A2M4BDU5 A0A2M3Z2J9 A0A2M3Z2K2 A0A1Q3G435 A0A1Q3G473 Q7QK77 A0A1Q3G4A2 F5HIV0 T1PEP9 A0A1I8M8M8 T1PLU1 A0A1J1IF34 T1PP38 A0A2J7R3N4 T1PLT6 W8B062 U5ER34 A0A1L8E3H2 A0A1L8E3V1 W8BF58 A0A2M4CVB8 B3LYM0 A0A034VFQ6 A0A2M4DLP2 A0A1I8M8M3 A0A0P8YMD8 A0A1I8M8N1 A0A0P8XZK3 A0A1I8M8M5 A0A0P9AR17 A0A0P8YF07 A0A0K8UZB7 B4K4U9 A0A034VE53 A0A0Q9XBK3 A0A0Q9X1I1 A0A0R3NJ46 A0A0R3NIZ5 A0A1I8M8N3 I5APG6 A0A224X707 A0A0Q9X1I8 A0A1W4VLK6 A0A1W4VK86 A0A1W4V8H5 A0A0M5J046 A0A1W4VLM1 A0A1W4V833 A0A0R3NP71 B4K4V0 A0A0R3NJ76 I5APG7 A0A0Q9X257 A0A0Q9XE49 B3P6F8 A0A1I8M8N4
PDB
3M0C
E-value=3.40276e-129,
Score=1184
Ontologies
GO
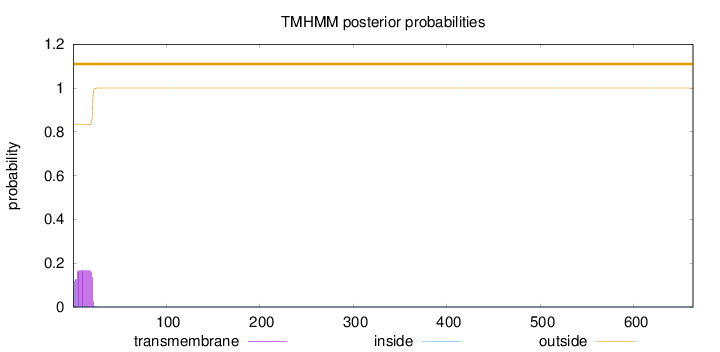
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.999172

Length:
663
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.19962
Exp number, first 60 AAs:
3.19944
Total prob of N-in:
0.16763
outside
1 - 663
Population Genetic Test Statistics
Pi
247.87627
Theta
182.398373
Tajima's D
1.388563
CLR
0.422838
CSRT
0.76121193940303
Interpretation
Uncertain
