Gene
KWMTBOMO02614
Pre Gene Modal
BGIBMGA002632
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Full name
Queuine tRNA-ribosyltransferase accessory subunit 2
Alternative Name
Queuine tRNA-ribosyltransferase domain-containing protein 1
Location in the cell
Mitochondrial Reliability : 2.06 Nuclear Reliability : 1.829
Sequence
CDS
ATGCAAGAATACAACATTCACATTCTTGGATTGAGTGAGACGAGATGGAACGGTTTTGGAGAAACGCAATGTGATTTTGGATACTCGTTATTGTACAGTGGTAAGGAAGAGGATGCGACCCGAGAGTATGGCGTGGGTTTGCTGCTAAGTCGCAGCGCTAAAAGTAGCTTACTCTCTTGGAAGCCAGTCTCGGAACGAATAATATGCGCCAGATACGGTCACGACTCGACACGAGTACGGAAGGTGACAGTGATACAATGCTACGCTCCCACTAACTCTGCTTCGACCGATGTAAAGTCACAATTCTACGAACAGCTGGGGCATACACTCAAAGGCATCAAGAAACAAGATATCGTGATTCTCATGGGCGACATAAATGCGAAGATGGGAAATGACAATGTAGACCGCGAATATTACATGGGACAATATGGGGCAACCGGAACTATTAACGAAAACGGAGAACTTTTCGGTGATCTATGTTCCACACATGGTCTAGTTATTGGAGGTACCCTATTTAAGCATAAGGAATGTCATAAAGTTACTTGGATATCACCGGACGGAAAGACAAAGAACCAAATCGATCACGTAGCTATCAGTAAGACATGGCGCTCTTCTCTTCTTGATGTGCGAAATAAGCGCGGCGCAGACATTGACAGTGATCACCATCTGGTGGTAGCAGAAGTTCGTCTTAAAATAGCAGTTCCCCACAGAAAGCCAGATAAAGCAGCTAGAAAATTTTATATCCGAAAACTGGAAGATAGCAATTGCCGAGAGAAGTTCAGCTTATCTCTCAAAAATAGTTTCGAGCTGCTTAAGGAAATAGAGGATGAGCAGATGGACGAGACGTGGGACCGCATACGTGGTGCTATAATAAAGCTGCTAAGGAAGTATTGGGGTATACCAATACAAAGCAAGAACCTTGGATATGCTATGTTGCGTCGGGAATACTCATCCACGCGATTAGTAATTAGAAAGCTGGCTAGGAGAGATCAGCGAAATGCAATCGATGAGCTAGCAGACCGAGCAAAATTGGCAGCCAAGAGGCAAGATATGAAAGAGCTTTACGATGTCACCAAGCGGATAATAAACAAAAAACATCATCCACGGAAAAAACCACTGAAAGACTGCTCGGGTCAGCTACTATGTACCACGGAAGAGCAGCTCGTGCGCTGGACGGAGCACTTTACTAAGTTGCTGTCCAAATGTCAGTCCCAACCCACAGACGGAATATCAGTACCCGACTTGCCTTTGGATATACCTATTGCACCGCCCACCTTTAATGAAATAGTAAAGGCAATTAAACAGCTGAAACCAGGTAAGGCCCCCGGTATTGACAATATCAATGCCGAAATGCTGAAGGCTGATGCACCGAGATCTGCAAGCTTGCTATATCCGCTCATCGTAAGGATTTGGGAGACTGAGCAAATACCGTCTCAATGGAAGCAGGGACTACTCGTGAAAGTTCCGAAGAAGGGCGATTTGTCTTCTTGTGACAACTGGAGAGGCATTACACTTCTTCCTTGCGCAGCAAAGGTCTTGGCTAGATTACTTCTAAACCGGATGTCCAAAAAGATCACTGGAACTCTTCGGGAGGAGCAAGCAGGTTTCTTACCGGGCCGATCATGCACAGACCATACAAACACTCTTCGCATCCTCATAGAACAAAGTGTCGAATGGCAGACTGAAATGATTCTTACGTTTGTAGACTTTGAGAAAGCGTTTGACACTGTACAGTGGAGCAAAATGTGGACCTGCTTAAAACAAAGAGGTATACCAAATAAAATCATTGGTATAATCCAGGCCTTATACAGAGAACAGCTACTTTGA
Protein
MQEYNIHILGLSETRWNGFGETQCDFGYSLLYSGKEEDATREYGVGLLLSRSAKSSLLSWKPVSERIICARYGHDSTRVRKVTVIQCYAPTNSASTDVKSQFYEQLGHTLKGIKKQDIVILMGDINAKMGNDNVDREYYMGQYGATGTINENGELFGDLCSTHGLVIGGTLFKHKECHKVTWISPDGKTKNQIDHVAISKTWRSSLLDVRNKRGADIDSDHHLVVAEVRLKIAVPHRKPDKAARKFYIRKLEDSNCREKFSLSLKNSFELLKEIEDEQMDETWDRIRGAIIKLLRKYWGIPIQSKNLGYAMLRREYSSTRLVIRKLARRDQRNAIDELADRAKLAAKRQDMKELYDVTKRIINKKHHPRKKPLKDCSGQLLCTTEEQLVRWTEHFTKLLSKCQSQPTDGISVPDLPLDIPIAPPTFNEIVKAIKQLKPGKAPGIDNINAEMLKADAPRSASLLYPLIVRIWETEQIPSQWKQGLLVKVPKKGDLSSCDNWRGITLLPCAAKVLARLLLNRMSKKITGTLREEQAGFLPGRSCTDHTNTLRILIEQSVEWQTEMILTFVDFEKAFDTVQWSKMWTCLKQRGIPNKIIGIIQALYREQLL
Summary
Description
Non-catalytic subunit of the queuine tRNA-ribosyltransferase (TGT) that catalyzes the base-exchange of a guanine (G) residue with queuine (Q) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine).
Cofactor
Zn(2+)
Subunit
Heterodimer of a catalytic subunit and an accessory subunit.
Similarity
Belongs to the queuine tRNA-ribosyltransferase family. QTRT2 subfamily.
Uniprot
D7F173
H9IZE7
A0A146MF87
A0A1B0GLA1
A0A336MSP8
Q4QQE8
+ More
A0A3B1KH88 A0A2B4R8E3 A0A2B4RLZ4 C7C207 A0A183MMI0 A0A1S3ISC1 A0A2T7PJY8 W4XQE1 A0A183NCD4 W5JEC3 H3B4N3 A0A183MGB1 A0A183LTH6 A0A0B7BT32 A0A336M488 A0A2G8K194 A0A183MP16 A0A183MTT9 A0A183KHV6 A0A2T7NHE8 A0A183N5W2 A0A183LFM6 A0A183MS45 A0A2S2PT92 A0A023F0H0 A0A2W1BNH4 A0A183L9Y2 A0A183NCM3 A0A146KNZ7 A0A016VQL3 A0A2P8XC08 A0A023F0F3 A0A0B7BP96 A0A023EX67 A0A183MMG7 A0A183MYH2 A0A183MVQ1 A0A2B7ZXS2 A0A0K8SN56 A0A183ME56 A0A3B1KAX6 J9L6X2 A0A2P8ZLT6 A0A2P8Z488 A0A183K1M3 A0A060Q6A2 A0A2P8YWA2 A0A2S2QKG5 X1WTQ9 D7F174 A0A034VWX2 A0A2P8Y2S1 A0A183N3Q5 A0A023F0M0 D7F170 A0A0V0G8V1 D7F161 A0A1Y1JWU5 A0A2P8ZFZ0 A0A023GD30 J9LWM8 A0A183N4V7 A0A023EZ18 A0A183MRN1 A0A183L9X8 W4YIG0 A0A016TPK0 A0A023EZ68 Q8MWP9 A0A2P8Z313 H2YWZ5 A0A023EWL3 Q8T5G5 A0A069DWU6 A0A1B0DRK3 A0A069DWR4 A0A0U4HM41 A0A2P8ZA67 T1E1T8 D7F163 A0A183KM31 A0A183LK64 A0A069DWW0 A0A0R3Q7D3 J9KNQ8
A0A3B1KH88 A0A2B4R8E3 A0A2B4RLZ4 C7C207 A0A183MMI0 A0A1S3ISC1 A0A2T7PJY8 W4XQE1 A0A183NCD4 W5JEC3 H3B4N3 A0A183MGB1 A0A183LTH6 A0A0B7BT32 A0A336M488 A0A2G8K194 A0A183MP16 A0A183MTT9 A0A183KHV6 A0A2T7NHE8 A0A183N5W2 A0A183LFM6 A0A183MS45 A0A2S2PT92 A0A023F0H0 A0A2W1BNH4 A0A183L9Y2 A0A183NCM3 A0A146KNZ7 A0A016VQL3 A0A2P8XC08 A0A023F0F3 A0A0B7BP96 A0A023EX67 A0A183MMG7 A0A183MYH2 A0A183MVQ1 A0A2B7ZXS2 A0A0K8SN56 A0A183ME56 A0A3B1KAX6 J9L6X2 A0A2P8ZLT6 A0A2P8Z488 A0A183K1M3 A0A060Q6A2 A0A2P8YWA2 A0A2S2QKG5 X1WTQ9 D7F174 A0A034VWX2 A0A2P8Y2S1 A0A183N3Q5 A0A023F0M0 D7F170 A0A0V0G8V1 D7F161 A0A1Y1JWU5 A0A2P8ZFZ0 A0A023GD30 J9LWM8 A0A183N4V7 A0A023EZ18 A0A183MRN1 A0A183L9X8 W4YIG0 A0A016TPK0 A0A023EZ68 Q8MWP9 A0A2P8Z313 H2YWZ5 A0A023EWL3 Q8T5G5 A0A069DWU6 A0A1B0DRK3 A0A069DWR4 A0A0U4HM41 A0A2P8ZA67 T1E1T8 D7F163 A0A183KM31 A0A183LK64 A0A069DWW0 A0A0R3Q7D3 J9KNQ8
Pubmed
EMBL
FJ265558
ADI61826.1
BABH01014180
BABH01014181
BABH01014182
BABH01014183
+ More
GDHC01000226 JAQ18403.1 AJWK01034673 UFQT01001780 SSX31783.1 BN000794 CAJ00236.1 LSMT01001244 PFX12648.1 LSMT01000478 PFX17357.1 FN356221 CAX83710.1 UZAI01017328 VDP23394.1 PZQS01000003 PVD33720.1 AAGJ04045270 UZAI01022169 VDP57417.1 ADMH02001629 ETN61698.1 AFYH01088828 UZAI01016872 VDP17428.1 UZAI01002797 VDO74729.1 HACG01049283 CEK96148.1 UFQS01000142 UFQT01000142 SSX00428.1 SSX20808.1 MRZV01000990 PIK41764.1 UZAI01017464 VDP25179.1 UZAI01017973 VDP31675.1 UZAK01036860 VDP56899.1 PZQS01000012 PVD20578.1 UZAI01019871 VDP48262.1 UZAI01000669 VDO55444.1 UZAI01017784 VDP29476.1 GGMR01020071 MBY32690.1 GBBI01004263 JAC14449.1 KZ150025 PZC74827.1 UZAI01000113 VDO48731.1 UZAI01022444 VDP57962.1 GDHC01021627 JAP97001.1 JARK01001341 EYC29874.1 PYGN01003402 PSN29545.1 GBBI01004261 JAC14451.1 HACG01048259 CEK95124.1 GAPW01000107 JAC13491.1 UZAI01017324 VDP23363.1 UZAI01018571 VDP38269.1 UZAI01018190 VDP34363.1 PDHN01000949 PGH37547.1 GBRD01011073 JAG54751.1 UZAI01016773 VDP15570.1 ABLF02018097 ABLF02043469 ABLF02056908 PYGN01000020 PSN57453.1 PYGN01000203 PSN51319.1 UZAK01032956 VDP33068.1 HE799691 CCH14900.1 PYGN01000324 PSN48495.1 GGMS01009011 MBY78214.1 ABLF02035454 ABLF02035459 ABLF02035461 ABLF02042121 FJ265559 ADI61827.1 GAKP01012607 JAC46345.1 PYGN01001004 PSN38555.1 UZAI01019418 VDP45194.1 GBBI01004198 JAC14514.1 FJ265555 ADI61823.1 GECL01001585 JAP04539.1 FJ265546 ADI61814.1 GEZM01098732 JAV53782.1 PYGN01000070 PSN55417.1 GBBM01003639 JAC31779.1 ABLF02042851 UZAI01019647 VDP46800.1 GBBI01004262 JAC14450.1 UZAI01017731 VDP28843.1 UZAI01000112 VDO48722.1 AAGJ04063063 AAGJ04063064 AAGJ04063065 JARK01001422 EYC04705.1 GBBI01004199 JAC14513.1 AF412214 AAM93546.1 PYGN01000219 PSN50892.1 GAPW01000120 JAC13478.1 AY027869 AAK14815.1 GBGD01000583 JAC88306.1 AJVK01001578 GBGD01000559 JAC88330.1 KU311054 ALX81665.1 PYGN01000129 PSN53384.1 GALA01001775 JAA93077.1 FJ265548 ADI61816.1 UZAK01038324 VDP60854.1 UZAI01001301 VDO60450.1 GBGD01000331 JAC88558.1 UZAG01001147 VDO10529.1 ABLF02039015
GDHC01000226 JAQ18403.1 AJWK01034673 UFQT01001780 SSX31783.1 BN000794 CAJ00236.1 LSMT01001244 PFX12648.1 LSMT01000478 PFX17357.1 FN356221 CAX83710.1 UZAI01017328 VDP23394.1 PZQS01000003 PVD33720.1 AAGJ04045270 UZAI01022169 VDP57417.1 ADMH02001629 ETN61698.1 AFYH01088828 UZAI01016872 VDP17428.1 UZAI01002797 VDO74729.1 HACG01049283 CEK96148.1 UFQS01000142 UFQT01000142 SSX00428.1 SSX20808.1 MRZV01000990 PIK41764.1 UZAI01017464 VDP25179.1 UZAI01017973 VDP31675.1 UZAK01036860 VDP56899.1 PZQS01000012 PVD20578.1 UZAI01019871 VDP48262.1 UZAI01000669 VDO55444.1 UZAI01017784 VDP29476.1 GGMR01020071 MBY32690.1 GBBI01004263 JAC14449.1 KZ150025 PZC74827.1 UZAI01000113 VDO48731.1 UZAI01022444 VDP57962.1 GDHC01021627 JAP97001.1 JARK01001341 EYC29874.1 PYGN01003402 PSN29545.1 GBBI01004261 JAC14451.1 HACG01048259 CEK95124.1 GAPW01000107 JAC13491.1 UZAI01017324 VDP23363.1 UZAI01018571 VDP38269.1 UZAI01018190 VDP34363.1 PDHN01000949 PGH37547.1 GBRD01011073 JAG54751.1 UZAI01016773 VDP15570.1 ABLF02018097 ABLF02043469 ABLF02056908 PYGN01000020 PSN57453.1 PYGN01000203 PSN51319.1 UZAK01032956 VDP33068.1 HE799691 CCH14900.1 PYGN01000324 PSN48495.1 GGMS01009011 MBY78214.1 ABLF02035454 ABLF02035459 ABLF02035461 ABLF02042121 FJ265559 ADI61827.1 GAKP01012607 JAC46345.1 PYGN01001004 PSN38555.1 UZAI01019418 VDP45194.1 GBBI01004198 JAC14514.1 FJ265555 ADI61823.1 GECL01001585 JAP04539.1 FJ265546 ADI61814.1 GEZM01098732 JAV53782.1 PYGN01000070 PSN55417.1 GBBM01003639 JAC31779.1 ABLF02042851 UZAI01019647 VDP46800.1 GBBI01004262 JAC14450.1 UZAI01017731 VDP28843.1 UZAI01000112 VDO48722.1 AAGJ04063063 AAGJ04063064 AAGJ04063065 JARK01001422 EYC04705.1 GBBI01004199 JAC14513.1 AF412214 AAM93546.1 PYGN01000219 PSN50892.1 GAPW01000120 JAC13478.1 AY027869 AAK14815.1 GBGD01000583 JAC88306.1 AJVK01001578 GBGD01000559 JAC88330.1 KU311054 ALX81665.1 PYGN01000129 PSN53384.1 GALA01001775 JAA93077.1 FJ265548 ADI61816.1 UZAK01038324 VDP60854.1 UZAI01001301 VDO60450.1 GBGD01000331 JAC88558.1 UZAG01001147 VDO10529.1 ABLF02039015
Proteomes
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR027124 Swc5/CFDP2
IPR036511 TGT-like_sf
IPR028592 QTRTD1
IPR002616 tRNA_ribo_trans-like
IPR005135 Endo/exonuclease/phosphatase
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR000637 HMGI/Y_DNA-bd_CS
IPR036691 Endo/exonu/phosph_ase_sf
IPR027124 Swc5/CFDP2
IPR036511 TGT-like_sf
IPR028592 QTRTD1
IPR002616 tRNA_ribo_trans-like
IPR005135 Endo/exonuclease/phosphatase
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR000637 HMGI/Y_DNA-bd_CS
Gene 3D
ProteinModelPortal
D7F173
H9IZE7
A0A146MF87
A0A1B0GLA1
A0A336MSP8
Q4QQE8
+ More
A0A3B1KH88 A0A2B4R8E3 A0A2B4RLZ4 C7C207 A0A183MMI0 A0A1S3ISC1 A0A2T7PJY8 W4XQE1 A0A183NCD4 W5JEC3 H3B4N3 A0A183MGB1 A0A183LTH6 A0A0B7BT32 A0A336M488 A0A2G8K194 A0A183MP16 A0A183MTT9 A0A183KHV6 A0A2T7NHE8 A0A183N5W2 A0A183LFM6 A0A183MS45 A0A2S2PT92 A0A023F0H0 A0A2W1BNH4 A0A183L9Y2 A0A183NCM3 A0A146KNZ7 A0A016VQL3 A0A2P8XC08 A0A023F0F3 A0A0B7BP96 A0A023EX67 A0A183MMG7 A0A183MYH2 A0A183MVQ1 A0A2B7ZXS2 A0A0K8SN56 A0A183ME56 A0A3B1KAX6 J9L6X2 A0A2P8ZLT6 A0A2P8Z488 A0A183K1M3 A0A060Q6A2 A0A2P8YWA2 A0A2S2QKG5 X1WTQ9 D7F174 A0A034VWX2 A0A2P8Y2S1 A0A183N3Q5 A0A023F0M0 D7F170 A0A0V0G8V1 D7F161 A0A1Y1JWU5 A0A2P8ZFZ0 A0A023GD30 J9LWM8 A0A183N4V7 A0A023EZ18 A0A183MRN1 A0A183L9X8 W4YIG0 A0A016TPK0 A0A023EZ68 Q8MWP9 A0A2P8Z313 H2YWZ5 A0A023EWL3 Q8T5G5 A0A069DWU6 A0A1B0DRK3 A0A069DWR4 A0A0U4HM41 A0A2P8ZA67 T1E1T8 D7F163 A0A183KM31 A0A183LK64 A0A069DWW0 A0A0R3Q7D3 J9KNQ8
A0A3B1KH88 A0A2B4R8E3 A0A2B4RLZ4 C7C207 A0A183MMI0 A0A1S3ISC1 A0A2T7PJY8 W4XQE1 A0A183NCD4 W5JEC3 H3B4N3 A0A183MGB1 A0A183LTH6 A0A0B7BT32 A0A336M488 A0A2G8K194 A0A183MP16 A0A183MTT9 A0A183KHV6 A0A2T7NHE8 A0A183N5W2 A0A183LFM6 A0A183MS45 A0A2S2PT92 A0A023F0H0 A0A2W1BNH4 A0A183L9Y2 A0A183NCM3 A0A146KNZ7 A0A016VQL3 A0A2P8XC08 A0A023F0F3 A0A0B7BP96 A0A023EX67 A0A183MMG7 A0A183MYH2 A0A183MVQ1 A0A2B7ZXS2 A0A0K8SN56 A0A183ME56 A0A3B1KAX6 J9L6X2 A0A2P8ZLT6 A0A2P8Z488 A0A183K1M3 A0A060Q6A2 A0A2P8YWA2 A0A2S2QKG5 X1WTQ9 D7F174 A0A034VWX2 A0A2P8Y2S1 A0A183N3Q5 A0A023F0M0 D7F170 A0A0V0G8V1 D7F161 A0A1Y1JWU5 A0A2P8ZFZ0 A0A023GD30 J9LWM8 A0A183N4V7 A0A023EZ18 A0A183MRN1 A0A183L9X8 W4YIG0 A0A016TPK0 A0A023EZ68 Q8MWP9 A0A2P8Z313 H2YWZ5 A0A023EWL3 Q8T5G5 A0A069DWU6 A0A1B0DRK3 A0A069DWR4 A0A0U4HM41 A0A2P8ZA67 T1E1T8 D7F163 A0A183KM31 A0A183LK64 A0A069DWW0 A0A0R3Q7D3 J9KNQ8
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
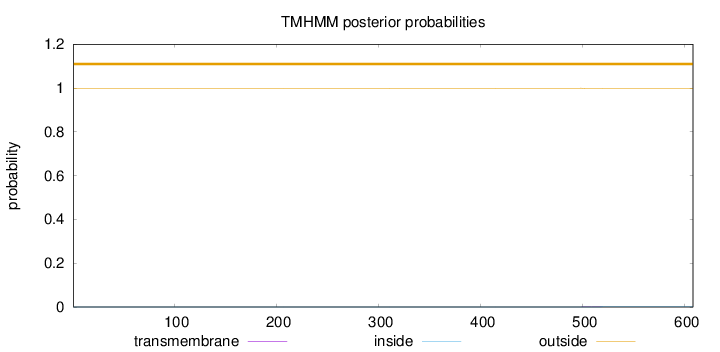
Length:
608
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00045
outside
1 - 608
Population Genetic Test Statistics
Pi
576.012105
Theta
239.798045
Tajima's D
4.416139
CLR
0.596177
CSRT
0.999850007499625
Interpretation
Possibly Balancing Selection
