Gene
KWMTBOMO02604
Pre Gene Modal
BGIBMGA002665
Annotation
PREDICTED:_adenylate_cyclase_type_7-like_[Papilio_machaon]
Full name
Adenylate cyclase
+ More
Guanylate cyclase
Adenylyl cyclase X E
Guanylate cyclase
Adenylyl cyclase X E
Location in the cell
PlasmaMembrane Reliability : 3.889
Sequence
CDS
ATGGGCTCTGTGGTGCTTCAAGTAGGCGCCTTAGAGCTCTTAGCAAGGTATTCAACGAACTCTGGGAGAGCGACCAGCGTTCTCTCATCATTACATGAATTCAGAACACAAGAAGTTGAATTGGCTGGTGGGAAACAGTGGAATTGGAAGTATTTAAGGGATCAATTTGATCAAAAAGATTTAGAAGGACTCTACAGAAAATACGATGACAAACTGAGAGATGTATTAATTCATATTTATGTATCTCTTCTAGTATTCTTTACGACTGTACACATCGTTTTAGTTGTGATATCTACATTTTCAGAGCAAATACAGTCAGAGTCAACATACTTAACAATGGGAATGTACGTTTTGCGTATTGCAATACCAGCTCTATCACTTTGCAAGAAGGTATATGAGCCCTTGAAGAAGAAGTGCAATTGGATGCCGATATTTATTACATTTTTTGTTACTTTGCACTTAGTTTTAACAGATATCGCTGTGTGCATCTACAGTTCGTTCGAGGGGATATCGTTGAGACCAGCATACGCAACGATGGCTTTGCTGTCTATTTACATATTTTTACCTATGAGAAACAATTTCCTGGTAATAATACTGGGTGTTTTGGTCAGCGCTATCTATGTACTGATCTTTGCTTTCTTCACTTATCGGCAGAACCCACAGATCGGTGTTGTCGTAGCATCAGATATAATTTATCTGATCGGCGTAAATCTAATGGGCGTGTACTTTAGGCTAATGAACGAAATAATTACAAGACGGTCCTTTTTGGACAGAAGAGCCTGTGTGGAGAGTACGTTAAGATTGAAATTCGTTAAGGATCAAGAAGAACGGCTGATGATGAGTATTTTACCAGAGCACATCGTATCCAAAGTAAGACAGGACATTAGAAATATGTTTTTGGGACTACACACTCGCGACTTAAGTTACACTATGAGATCATTCAATCAGTTATACGTCGAAGAACACGAAAATGTCAGTATTCTTTATGCGGATGTCGTTAATTACACAATGATATCAACAAGTTTATCTCCCATGAGAATGGTGGAATTACTGAACGAGCTTTTCGGAAGATTTGACGAGGCTTCAGAGGAATACGATGTACTACGGATAAAATTCCTTGGTGACTGTTACTACTGCGTCAGTGGGATTCCAAAGCCTTCTGTTTATCATGCGAAAAACTGCGTTGATTTAGGACTGGAAATGATTCATATCATCAAAGATGTCAGAGAAAAACGCTCCTTAAACATAGATATGAGGATAGGTGTACATTCCGGGAGAATTCTGAGTGGCTTGATAGGTATCAGAAAATGGCAATACGATGTTTGGTCAAAAGACGTTACCATAGCAAACAAAATGGAATCCACCGGAAAAGCTGGAAAAGTTCACGTAACCAAGCAAACGCTAGAACTCCTTATAGATTTTGCAAGAGAATATATTATTGAACCAAATTTTGAATCTCAAAATGATCCTTTCATCACAAAAAATAAACTGGAAACCTTTCTTCTGTCACGACCAGCCAGGCCTCCAGAATACAAACCCTTCCGTAGAGCATCCGTTGGATTTAACAAGATAATTACAACTAAAACTTCAACATCTAGACGATCGGATAACAGAAATTCAACGAGACGAACAACAACGTTCATGGATGAGAACTTGGTGGAATACCAGCAAATGTTGAAAGCTGCTGACGCGCAGATGGCAAAGGAGATCGAAGAAATGACAAAGGGAAAAGAATTTTTCAAGAAAGAGTCAAACTTGAACCGTTGTTCGCTAATGTTCAAAGACTTCCGTCTTGAAAAGATGTTCCTTTTGCTGTCGGATCCACTATTTAAATATTACATAACATGCTGTCTTATTATTTTATGCTTGATTTTATTGATCAATGGACTAACTACAAATTGGCTTTATGACTTCCATTGGATTACTTGGCTGCTTTTTGGATTCCTTGTTATCGGACTAATTATAATGCAACCCTTCACGTGGTTCCACTTTTTATGGACAAAATACAAAGGAATCGATGAACCGAAAAACAAATATTTGCGATATATATATGACATTTCTTCAAATATAATAAGATCTGCTAAAATAAGAACAATAATGTATTTATTGATAAGTCTAGGTCTGGCTGCGACTTCAGTAATTACAGTTGTTGGATGTTCTGTTGTAGAAGTAGAATTAGTGGATGCTGAATCAGTATTATCCAATTGTGTATCGTCATGGCACGTCACACAATGTTGGGGGCTGGCGCTCCTTTTGAATTTCCTATTCAGCAGAGTTTTTTTCATATTTAAGTGGATCGTTGCCGGTGGAATGACAGTGTTCTACCTATGGGTTGTTTGGAAAATAAAAACTTCTCTTTTTGCTGAAGATGCAACATGGAATGTTGGAACAACTGAACACAGAAACCGATTAGACCATTTATGGCAATTGCAACTAAGTGAGGAGCAACAAGAAGCTGAAACTATGCTTAAAGTTAATAATATGCTTTTGGAGAATATTTTACCTGCTCATGTAGTTCAGGTGTACTTAAATTTGAATAGGTCAATAGATGAGTTGTATTATGAAAAGTATGACAATGTGGCCGTAATGTTCGCCTCCCTCACTGACTACAAACTGGGCCTAGAAGGTGACGCAGATATGAGTGATAAATTTGTGTTAAGCATATTAGATGAAGTAATTTCAGATTTCGACCGGTTGCTCCTAACAGGAACATCGGCTCATAGAGTTGAAAAAATTAAAATGGCAGGTTGGACTTATATGGCTGCATGCGGATTAGATCCAAATAGACGCGAATCACCCGATGCTATGTCTCATACATCTAACAGTCCAAATAACAGTCCATATAGTAAATCCGTTGTATTAACCATGGTTGAATTCGCTGCAGCTATGATGATGCAACTTCAAAATTTTAATCGAGGGTCATTTCAAAGCTTTGAAGGACTCAATTTAAGAATAGGTATCTCACACGGCGAAGTAGCAGCGGGCGTGGTGGGGTCTTTAAAGCCGCTTTATGATATTTGGGGACATGCAGTAAACATGGCTTCAAGAATGGATTCGACTGGACAAACTGGTAAAATTCAAGTTACCGAAAATACAGCAATATTACTGGACGAATGTGACATTCTTACGACTTACCGAGGAGAGACTTTCGTCAAGGGAGCAGGATACATCAAAACATATTATGTTCCACTTGATGAAAATTTCAATTTAATAAAAAAGGATCAAAGTCTACATTACTATAGATTAGAAGATAGGAATCGTTATTATAACCTTAGGAATACGAACTTTGAAGAACCTTATTCAGATAATGACAATATTGAATCAGAAACTTACGATTCTACTTTTAGTGTTAGAGATCTTAGTATTCTTAGTGACAATTCTAGTCTATACGTCGAAGAACACGAAAATGTCAGCATTCTTTATGCGGATGTCGTTAATTACACAATGATATCAACAAGTTTGTCTCCCATGAGAATGGTGGAATTACTGAACGAGCTTTTCGTAAGATTTGACGAGGCTTCAGAGGAATACGATGTACTACAAATAAAATTCCTTGGCGACTGTTACTACTGCGTCAGTGGGATTCCAAAGCCTTCTGTTTATCATGCGAAAAACTGCGTTGATTTAGGACTGGAAATGATTCATATCATCAAAGATGTCAGAGAAAAACGCTCCTTAAACATAGATATGAGGATAGGTGTACATTCCGGGAGAATTCTGAGTGGCTTAATAGGTATCAGAAAATGGCAATACGATGTTTGGTCAAAAGACGTTACCATAGCAAACAAAATGGAATCCACCGGAAAAGCTGGAAAAGTTCACGTAACCAAGCAAACGCTAGAACTCCTTATAGATTTTGCAAGAGAATATATTATTGAACCAAATTTTGAATCTCAAAATGATCCTTTCATCACAAAAAATAAATTGGAAACCTTTCTTCTGTCACGACCAGCCAGACCTCCAGAATACAAACCCTTCCGTAGAGCATCCGTTGGATTTAACAAGATAATTACAACTAAAACTTCAACGTCTAGACGATCGGATAACAGAAATTCAACGAGACGAACAACAACGTTCATGGACGAGAACTTGGTGGAATATCAGCAAATGTTGAAGGCTGCTGACGCGCAGATGGCGAAGGAGATCGAAGAAATGACAAAGGGAAAAGAATTTTTCAAGAAAGAATCAAACTTGAACCGATGTTCGCTAATGTTCAAAGACTTCCGTCTTGAAAAAATGTTCCTTTTGCTGTCGGACCCTCTATTTAAATATTACATAACATGCTGTCTTATTATTTTATGCTTGATTTTATTGATCAATGGATTAACTACAAATTGGCTTTATGATTTCCATTGGGTGTACTTAAATTTGAATAGGTCAATAGACGAGTTGTATTATGAAAAGTATGACAATGTGGCAGTAATGTTCGCCTCCCTCACTGACTACAAACTGGGCCTAGAAGGTGACGCAGATATGAGTGATAAATTTGTGTTAAGCATATTAGATGAAGTAATTTCAGATTTCGACCGGTTGCTCCTAACAGGAACATCGGCTCATAGAGTTGAAAAAATTAAAATGGCAGGTTGGACTTATATGGCTGCATGCGGATTAGATCCAAATAGACGCGAATCACCAGATGCTATGTCTCGTAACCATGGTTGA
Protein
MGSVVLQVGALELLARYSTNSGRATSVLSSLHEFRTQEVELAGGKQWNWKYLRDQFDQKDLEGLYRKYDDKLRDVLIHIYVSLLVFFTTVHIVLVVISTFSEQIQSESTYLTMGMYVLRIAIPALSLCKKVYEPLKKKCNWMPIFITFFVTLHLVLTDIAVCIYSSFEGISLRPAYATMALLSIYIFLPMRNNFLVIILGVLVSAIYVLIFAFFTYRQNPQIGVVVASDIIYLIGVNLMGVYFRLMNEIITRRSFLDRRACVESTLRLKFVKDQEERLMMSILPEHIVSKVRQDIRNMFLGLHTRDLSYTMRSFNQLYVEEHENVSILYADVVNYTMISTSLSPMRMVELLNELFGRFDEASEEYDVLRIKFLGDCYYCVSGIPKPSVYHAKNCVDLGLEMIHIIKDVREKRSLNIDMRIGVHSGRILSGLIGIRKWQYDVWSKDVTIANKMESTGKAGKVHVTKQTLELLIDFAREYIIEPNFESQNDPFITKNKLETFLLSRPARPPEYKPFRRASVGFNKIITTKTSTSRRSDNRNSTRRTTTFMDENLVEYQQMLKAADAQMAKEIEEMTKGKEFFKKESNLNRCSLMFKDFRLEKMFLLLSDPLFKYYITCCLIILCLILLINGLTTNWLYDFHWITWLLFGFLVIGLIIMQPFTWFHFLWTKYKGIDEPKNKYLRYIYDISSNIIRSAKIRTIMYLLISLGLAATSVITVVGCSVVEVELVDAESVLSNCVSSWHVTQCWGLALLLNFLFSRVFFIFKWIVAGGMTVFYLWVVWKIKTSLFAEDATWNVGTTEHRNRLDHLWQLQLSEEQQEAETMLKVNNMLLENILPAHVVQVYLNLNRSIDELYYEKYDNVAVMFASLTDYKLGLEGDADMSDKFVLSILDEVISDFDRLLLTGTSAHRVEKIKMAGWTYMAACGLDPNRRESPDAMSHTSNSPNNSPYSKSVVLTMVEFAAAMMMQLQNFNRGSFQSFEGLNLRIGISHGEVAAGVVGSLKPLYDIWGHAVNMASRMDSTGQTGKIQVTENTAILLDECDILTTYRGETFVKGAGYIKTYYVPLDENFNLIKKDQSLHYYRLEDRNRYYNLRNTNFEEPYSDNDNIESETYDSTFSVRDLSILSDNSSLYVEEHENVSILYADVVNYTMISTSLSPMRMVELLNELFVRFDEASEEYDVLQIKFLGDCYYCVSGIPKPSVYHAKNCVDLGLEMIHIIKDVREKRSLNIDMRIGVHSGRILSGLIGIRKWQYDVWSKDVTIANKMESTGKAGKVHVTKQTLELLIDFAREYIIEPNFESQNDPFITKNKLETFLLSRPARPPEYKPFRRASVGFNKIITTKTSTSRRSDNRNSTRRTTTFMDENLVEYQQMLKAADAQMAKEIEEMTKGKEFFKKESNLNRCSLMFKDFRLEKMFLLLSDPLFKYYITCCLIILCLILLINGLTTNWLYDFHWVYLNLNRSIDELYYEKYDNVAVMFASLTDYKLGLEGDADMSDKFVLSILDEVISDFDRLLLTGTSAHRVEKIKMAGWTYMAACGLDPNRRESPDAMSRNHG
Summary
Description
Catalyzes the formation of the signaling molecule cAMP in response to G-protein signaling.
Catalytic Activity
ATP = 3',5'-cyclic AMP + diphosphate
GTP = 3',5'-cyclic GMP + diphosphate
GTP = 3',5'-cyclic GMP + diphosphate
Cofactor
Mg(2+)
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
ATP-binding
Complete proteome
Lyase
Magnesium
Membrane
Metal-binding
Nucleotide-binding
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Adenylyl cyclase X E
Uniprot
A0A2A4JWP0
A0A1A9VJ86
E2C8S2
A0A088A9P4
A0A158NDG4
A0A151I8P7
+ More
A0A0L7QXT2 A0A151WG64 A0A195DJ43 A0A195B1A1 E1ZWP3 K7IW63 A0A195FI12 A0A026WVL9 A0A232F4A6 Q17AS9 A0A1Q3FH84 A0A182QY05 B0W3P0 A0A182JM26 A0A182NHL1 A0A182X6E4 A0A182WIR8 A0A182JNX9 A0A182P3J4 A0A182KRT2 A0A182VBB7 A0A1Q3FH75 W5JPE3 A0A182XZ95 A0A182FFX0 Q7QJS0 A0A1B0D5W6 A0A182I3Q0 A0A182RFN9 A0A0A1XK61 A0A1L8E010 A0A0K8VG24 A0A0K8VJ34 A0A1I8P6M3 W8BSK9 A0A1L8DZR4 A0A1I8P6L0 A0A1I8MAK7 A0A1B0BR96 A0A1A9XV14 A0A1A9X0W6 A0A084VFC6 A0A1B0G1W6 A0A0M4EKS3 A0A1W4W4V1 B3M753 Q9W038 Q8IGU9 Q9NJ99 B3NBF6 B4PEC4 A0A0J9RMB5 A0A0P8XSB8 W8C4S9 A0A0R1DUK7 B4QN42 B4J0I9 A0A3B0KP39 B4H5T3 A0A0Q9WUX1 Q2LYT8 B4LBX0 A0A0M5IYV0 B3N3B7 Q9VK50 B3MNI0
A0A0L7QXT2 A0A151WG64 A0A195DJ43 A0A195B1A1 E1ZWP3 K7IW63 A0A195FI12 A0A026WVL9 A0A232F4A6 Q17AS9 A0A1Q3FH84 A0A182QY05 B0W3P0 A0A182JM26 A0A182NHL1 A0A182X6E4 A0A182WIR8 A0A182JNX9 A0A182P3J4 A0A182KRT2 A0A182VBB7 A0A1Q3FH75 W5JPE3 A0A182XZ95 A0A182FFX0 Q7QJS0 A0A1B0D5W6 A0A182I3Q0 A0A182RFN9 A0A0A1XK61 A0A1L8E010 A0A0K8VG24 A0A0K8VJ34 A0A1I8P6M3 W8BSK9 A0A1L8DZR4 A0A1I8P6L0 A0A1I8MAK7 A0A1B0BR96 A0A1A9XV14 A0A1A9X0W6 A0A084VFC6 A0A1B0G1W6 A0A0M4EKS3 A0A1W4W4V1 B3M753 Q9W038 Q8IGU9 Q9NJ99 B3NBF6 B4PEC4 A0A0J9RMB5 A0A0P8XSB8 W8C4S9 A0A0R1DUK7 B4QN42 B4J0I9 A0A3B0KP39 B4H5T3 A0A0Q9WUX1 Q2LYT8 B4LBX0 A0A0M5IYV0 B3N3B7 Q9VK50 B3MNI0
EC Number
4.6.1.1
4.6.1.2
4.6.1.2
Pubmed
EMBL
NWSH01000437
PCG76437.1
GL453694
EFN75639.1
ADTU01012487
ADTU01012488
+ More
KQ978338 KYM95041.1 KQ414699 KOC63427.1 KQ983185 KYQ46836.1 KQ980800 KYN12900.1 KQ976690 KYM78065.1 GL434869 EFN74401.1 KQ981560 KYN39907.1 KK107083 EZA60062.1 NNAY01001046 OXU25359.1 CH477330 EAT43379.1 GFDL01008129 JAV26916.1 AXCN02000593 DS231833 EDS32187.1 GFDL01008124 JAV26921.1 ADMH02000483 ETN66262.1 AAAB01008807 EAA04045.4 AJVK01000420 APCN01000217 GBXI01002901 JAD11391.1 GFDF01002098 JAV11986.1 GDHF01014526 JAI37788.1 GDHF01013415 JAI38899.1 GAMC01004433 JAC02123.1 GFDF01002106 JAV11978.1 JXJN01019007 ATLV01012374 KE524785 KFB36670.1 CCAG010004392 CP012525 ALC43910.1 CH902618 EDV38714.1 AE014296 BT053678 AAF47621.2 ACK77593.1 BT001587 AAN71342.1 AF177931 AAF33110.1 CH954178 EDV50278.1 CM000159 EDW92962.1 CM002912 KMY97061.1 KMY97063.1 KPU77597.1 GAMC01002192 JAC04364.1 KRK00840.1 CM000363 EDX08924.1 CH916366 EDV96825.1 OUUW01000012 SPP87636.1 CH479212 EDW33150.1 CH940647 KRF84555.1 KRF84556.1 CH379069 EAL29772.2 EDW69770.2 ALC44268.1 CH954177 EDV58757.1 AE014134 BT126324 CH902620 EDV32088.2
KQ978338 KYM95041.1 KQ414699 KOC63427.1 KQ983185 KYQ46836.1 KQ980800 KYN12900.1 KQ976690 KYM78065.1 GL434869 EFN74401.1 KQ981560 KYN39907.1 KK107083 EZA60062.1 NNAY01001046 OXU25359.1 CH477330 EAT43379.1 GFDL01008129 JAV26916.1 AXCN02000593 DS231833 EDS32187.1 GFDL01008124 JAV26921.1 ADMH02000483 ETN66262.1 AAAB01008807 EAA04045.4 AJVK01000420 APCN01000217 GBXI01002901 JAD11391.1 GFDF01002098 JAV11986.1 GDHF01014526 JAI37788.1 GDHF01013415 JAI38899.1 GAMC01004433 JAC02123.1 GFDF01002106 JAV11978.1 JXJN01019007 ATLV01012374 KE524785 KFB36670.1 CCAG010004392 CP012525 ALC43910.1 CH902618 EDV38714.1 AE014296 BT053678 AAF47621.2 ACK77593.1 BT001587 AAN71342.1 AF177931 AAF33110.1 CH954178 EDV50278.1 CM000159 EDW92962.1 CM002912 KMY97061.1 KMY97063.1 KPU77597.1 GAMC01002192 JAC04364.1 KRK00840.1 CM000363 EDX08924.1 CH916366 EDV96825.1 OUUW01000012 SPP87636.1 CH479212 EDW33150.1 CH940647 KRF84555.1 KRF84556.1 CH379069 EAL29772.2 EDW69770.2 ALC44268.1 CH954177 EDV58757.1 AE014134 BT126324 CH902620 EDV32088.2
Proteomes
UP000218220
UP000078200
UP000008237
UP000005203
UP000005205
UP000078542
+ More
UP000053825 UP000075809 UP000078492 UP000078540 UP000000311 UP000002358 UP000078541 UP000053097 UP000215335 UP000008820 UP000075886 UP000002320 UP000075880 UP000075884 UP000076407 UP000075920 UP000075881 UP000075885 UP000075882 UP000075903 UP000000673 UP000076408 UP000069272 UP000007062 UP000092462 UP000075840 UP000075900 UP000095300 UP000095301 UP000092460 UP000092443 UP000091820 UP000030765 UP000092444 UP000092553 UP000192221 UP000007801 UP000000803 UP000008711 UP000002282 UP000000304 UP000001070 UP000268350 UP000008744 UP000008792 UP000001819
UP000053825 UP000075809 UP000078492 UP000078540 UP000000311 UP000002358 UP000078541 UP000053097 UP000215335 UP000008820 UP000075886 UP000002320 UP000075880 UP000075884 UP000076407 UP000075920 UP000075881 UP000075885 UP000075882 UP000075903 UP000000673 UP000076408 UP000069272 UP000007062 UP000092462 UP000075840 UP000075900 UP000095300 UP000095301 UP000092460 UP000092443 UP000091820 UP000030765 UP000092444 UP000092553 UP000192221 UP000007801 UP000000803 UP000008711 UP000002282 UP000000304 UP000001070 UP000268350 UP000008744 UP000008792 UP000001819
Pfam
Interpro
IPR018297
A/G_cyclase_CS
+ More
IPR009398 Adcy_conserved_dom
IPR029787 Nucleotide_cyclase
IPR032628 AC_N
IPR001054 A/G_cyclase
IPR030672 Adcy
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR000742 EGF-like_dom
IPR008211 Laminin_N
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR038684 Laminin_N_sf
IPR000719 Prot_kinase_dom
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR009398 Adcy_conserved_dom
IPR029787 Nucleotide_cyclase
IPR032628 AC_N
IPR001054 A/G_cyclase
IPR030672 Adcy
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR000742 EGF-like_dom
IPR008211 Laminin_N
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR038684 Laminin_N_sf
IPR000719 Prot_kinase_dom
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
Gene 3D
ProteinModelPortal
A0A2A4JWP0
A0A1A9VJ86
E2C8S2
A0A088A9P4
A0A158NDG4
A0A151I8P7
+ More
A0A0L7QXT2 A0A151WG64 A0A195DJ43 A0A195B1A1 E1ZWP3 K7IW63 A0A195FI12 A0A026WVL9 A0A232F4A6 Q17AS9 A0A1Q3FH84 A0A182QY05 B0W3P0 A0A182JM26 A0A182NHL1 A0A182X6E4 A0A182WIR8 A0A182JNX9 A0A182P3J4 A0A182KRT2 A0A182VBB7 A0A1Q3FH75 W5JPE3 A0A182XZ95 A0A182FFX0 Q7QJS0 A0A1B0D5W6 A0A182I3Q0 A0A182RFN9 A0A0A1XK61 A0A1L8E010 A0A0K8VG24 A0A0K8VJ34 A0A1I8P6M3 W8BSK9 A0A1L8DZR4 A0A1I8P6L0 A0A1I8MAK7 A0A1B0BR96 A0A1A9XV14 A0A1A9X0W6 A0A084VFC6 A0A1B0G1W6 A0A0M4EKS3 A0A1W4W4V1 B3M753 Q9W038 Q8IGU9 Q9NJ99 B3NBF6 B4PEC4 A0A0J9RMB5 A0A0P8XSB8 W8C4S9 A0A0R1DUK7 B4QN42 B4J0I9 A0A3B0KP39 B4H5T3 A0A0Q9WUX1 Q2LYT8 B4LBX0 A0A0M5IYV0 B3N3B7 Q9VK50 B3MNI0
A0A0L7QXT2 A0A151WG64 A0A195DJ43 A0A195B1A1 E1ZWP3 K7IW63 A0A195FI12 A0A026WVL9 A0A232F4A6 Q17AS9 A0A1Q3FH84 A0A182QY05 B0W3P0 A0A182JM26 A0A182NHL1 A0A182X6E4 A0A182WIR8 A0A182JNX9 A0A182P3J4 A0A182KRT2 A0A182VBB7 A0A1Q3FH75 W5JPE3 A0A182XZ95 A0A182FFX0 Q7QJS0 A0A1B0D5W6 A0A182I3Q0 A0A182RFN9 A0A0A1XK61 A0A1L8E010 A0A0K8VG24 A0A0K8VJ34 A0A1I8P6M3 W8BSK9 A0A1L8DZR4 A0A1I8P6L0 A0A1I8MAK7 A0A1B0BR96 A0A1A9XV14 A0A1A9X0W6 A0A084VFC6 A0A1B0G1W6 A0A0M4EKS3 A0A1W4W4V1 B3M753 Q9W038 Q8IGU9 Q9NJ99 B3NBF6 B4PEC4 A0A0J9RMB5 A0A0P8XSB8 W8C4S9 A0A0R1DUK7 B4QN42 B4J0I9 A0A3B0KP39 B4H5T3 A0A0Q9WUX1 Q2LYT8 B4LBX0 A0A0M5IYV0 B3N3B7 Q9VK50 B3MNI0
PDB
3MAA
E-value=2.21167e-53,
Score=534
Ontologies
PATHWAY
GO
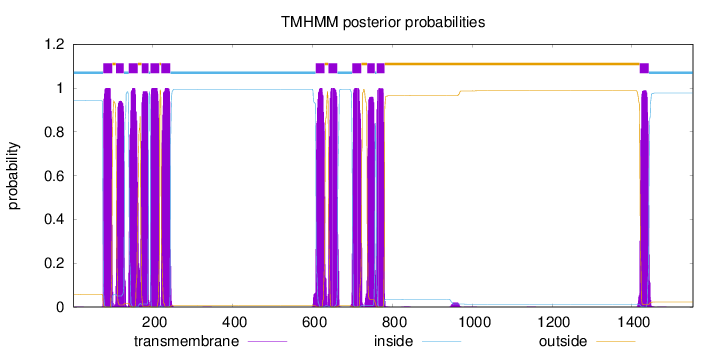
Topology
Subcellular location
Membrane
Length:
1553
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
256.288989999999
Exp number, first 60 AAs:
0.01294
Total prob of N-in:
0.94166
inside
1 - 75
TMhelix
76 - 98
outside
99 - 107
TMhelix
108 - 127
inside
128 - 139
TMhelix
140 - 162
outside
163 - 171
TMhelix
172 - 189
inside
190 - 193
TMhelix
194 - 216
outside
217 - 220
TMhelix
221 - 243
inside
244 - 607
TMhelix
608 - 630
outside
631 - 639
TMhelix
640 - 662
inside
663 - 699
TMhelix
700 - 722
outside
723 - 736
TMhelix
737 - 756
inside
757 - 760
TMhelix
761 - 780
outside
781 - 1419
TMhelix
1420 - 1442
inside
1443 - 1553
Population Genetic Test Statistics
Pi
221.331491
Theta
195.252862
Tajima's D
0.243118
CLR
0.493125
CSRT
0.44012799360032
Interpretation
Possibly Positive selection
