Gene
KWMTBOMO02603 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002630
Annotation
PREDICTED:_uncharacterized_protein_LOC105387556_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 2.837
Sequence
CDS
ATGTTAAAAGTGTTCGTCGTTGTCGTGTGCACGTTGGGCGCATCTCAGCTCTGCGCTGCGTTATACACGCAAAAGGTTGCAGTGTCCTTTCCTAAGGATAAAACGACAATAGTCGTGGAGGCCATGAAAAGCTGTATCGCGAAGACAGGAGCGAATCCGAACGTTATAGAAGTGATCAGTAGCGGCAAGGTATCTGAAGATGAAAAATTCAAAGAATTCTTTTACTGCGCTTGCAATGATATAGGAGTTGTAAATCCTGACGGACACATAAAGGTTAAAGAGTGTATTGAGCTCTTCCCAAAAGAAACACAGCCACTGGTTGAACCTGTCATTAAGAATTGTGACAAAGAAGGGGTCAATAAATACGATACACTTTTTAAATATTTAAAGTGCTTTCAAGAAACGTCTCCAGTTCGTGTTACGCTAGCATAA
Protein
MLKVFVVVVCTLGASQLCAALYTQKVAVSFPKDKTTIVVEAMKSCIAKTGANPNVIEVISSGKVSEDEKFKEFFYCACNDIGVVNPDGHIKVKECIELFPKETQPLVEPVIKNCDKEGVNKYDTLFKYLKCFQETSPVRVTLA
Summary
Uniprot
H9IZE5
A0A1E1W8T4
A0A345BER8
A0A2K8GKM7
A0A2K8GKL5
A0A2R4SBD9
+ More
A0A1B3P5J6 M9YYA4 A0A2K8GKN7 A0A0K8TUV6 C0SQ81 A0A0L7L7A8 A0A2K8GKQ9 H9IZE4 A0A076E7E0 A0A2H4G2B1 A0A076ED46 A0A1I9HZL2 A0A2K8GKN1 D6WS46 A0A1C8K2S9 A0A2R3ZV81 A0A310SBA3 D6WM81 G4Y3X2 A0A0X8RDP5 A0A1U9W501 D6WM83 A0A0S3J2L4 A0A1B4ZBJ7 A0A346TI01 A0A142FH84 U5YQF6 A0A1B1FKI3 A0A0T6BA30 A0A109NPF0 Q8WRW0 A0A1L2BL94 A0A0X8U7T9 A0A1W6QYA6 A0A1I8PZY0 D2D0C4 A0A1J0KKD4 A0A194PVX5 A0A1J0KKY1 B4KAZ8 A0A0C5BAY9 A0A1J0KKF9 A0A3G6V7G8 D4P5A8 A0A1I8N7H7 A0A0K8WJ37 A0A165UPF4 A0A3G2LEH8 W8W3V2 A0A194R1X8 F4Y5P3 A0A166IG76 F1CEY0 W8W3U3 A0A1S4FXS6 A0A075CKZ3 W8BUI1 B4L070 A0A0S3J2I5 B4MBS7 A0A386H995 A0A0X9EZ19
A0A1B3P5J6 M9YYA4 A0A2K8GKN7 A0A0K8TUV6 C0SQ81 A0A0L7L7A8 A0A2K8GKQ9 H9IZE4 A0A076E7E0 A0A2H4G2B1 A0A076ED46 A0A1I9HZL2 A0A2K8GKN1 D6WS46 A0A1C8K2S9 A0A2R3ZV81 A0A310SBA3 D6WM81 G4Y3X2 A0A0X8RDP5 A0A1U9W501 D6WM83 A0A0S3J2L4 A0A1B4ZBJ7 A0A346TI01 A0A142FH84 U5YQF6 A0A1B1FKI3 A0A0T6BA30 A0A109NPF0 Q8WRW0 A0A1L2BL94 A0A0X8U7T9 A0A1W6QYA6 A0A1I8PZY0 D2D0C4 A0A1J0KKD4 A0A194PVX5 A0A1J0KKY1 B4KAZ8 A0A0C5BAY9 A0A1J0KKF9 A0A3G6V7G8 D4P5A8 A0A1I8N7H7 A0A0K8WJ37 A0A165UPF4 A0A3G2LEH8 W8W3V2 A0A194R1X8 F4Y5P3 A0A166IG76 F1CEY0 W8W3U3 A0A1S4FXS6 A0A075CKZ3 W8BUI1 B4L070 A0A0S3J2I5 B4MBS7 A0A386H995 A0A0X9EZ19
Pubmed
EMBL
BABH01014228
BABH01014229
GDQN01007773
JAT83281.1
MG788224
AXF48742.1
+ More
KX585290 ARO70183.1 KX585285 ARO70178.1 MF066361 AVZ44708.1 KX655922 AOG12871.1 KC492498 KC763854 AGK24577.1 AGM38611.1 KX585289 ARO70182.1 GCVX01000157 JAI18073.1 AB353891 BAH36760.1 JTDY01002515 KOB71255.1 KX585311 ARO70204.1 KF487604 AII01002.1 KY130465 APG32533.1 KF487582 AII00980.1 KF977559 AIX97021.1 KX585287 ARO70180.1 KQ971354 EFA07547.1 KR780581 ANE37555.1 MH026104 AVR54528.1 GGOB01000085 MCH29481.1 KQ971343 EFA04687.2 JF937664 AEP27187.1 KJ186834 AIX97146.1 KX900465 AQY18977.1 EFA04593.1 KT381489 ALR72495.1 LC027689 BAV56792.1 MG751816 AXU25084.1 KT281925 AMQ76470.1 KF445134 KM251641 AGZ93682.1 AKC58522.1 KT268307 ANQ46501.1 LJIG01002781 KRT84159.1 KJ186814 AIX97126.1 AF393499 AAL60424.1 KT327213 ALS03856.1 KJ186818 AIX97130.1 KY056628 ARO46435.1 FJ233072 ACO83214.1 KX290616 APC94187.1 KQ459589 KPI97536.1 KX298775 APC94292.1 CH933806 EDW14676.1 KP071933 AJM71493.1 KX290639 APC94210.1 MH027968 AZB49386.1 GU575295 ADD70031.1 GDHF01001148 JAI51166.1 KU975055 AMY98994.1 MH937232 AYN70647.1 HG764550 CDJ79886.1 KQ460870 KPJ11697.1 GQ477025 ACZ58030.1 KT347569 ANA10230.1 HQ234489 ADY17884.1 HG764551 CDJ79887.1 KF460432 AHA39268.1 GAMC01003683 JAC02873.1 CH933809 EDW18016.1 KT381502 ALR72508.1 CH940656 EDW58548.1 MG546557 AYD42180.1 KP453838 ALM64969.1
KX585290 ARO70183.1 KX585285 ARO70178.1 MF066361 AVZ44708.1 KX655922 AOG12871.1 KC492498 KC763854 AGK24577.1 AGM38611.1 KX585289 ARO70182.1 GCVX01000157 JAI18073.1 AB353891 BAH36760.1 JTDY01002515 KOB71255.1 KX585311 ARO70204.1 KF487604 AII01002.1 KY130465 APG32533.1 KF487582 AII00980.1 KF977559 AIX97021.1 KX585287 ARO70180.1 KQ971354 EFA07547.1 KR780581 ANE37555.1 MH026104 AVR54528.1 GGOB01000085 MCH29481.1 KQ971343 EFA04687.2 JF937664 AEP27187.1 KJ186834 AIX97146.1 KX900465 AQY18977.1 EFA04593.1 KT381489 ALR72495.1 LC027689 BAV56792.1 MG751816 AXU25084.1 KT281925 AMQ76470.1 KF445134 KM251641 AGZ93682.1 AKC58522.1 KT268307 ANQ46501.1 LJIG01002781 KRT84159.1 KJ186814 AIX97126.1 AF393499 AAL60424.1 KT327213 ALS03856.1 KJ186818 AIX97130.1 KY056628 ARO46435.1 FJ233072 ACO83214.1 KX290616 APC94187.1 KQ459589 KPI97536.1 KX298775 APC94292.1 CH933806 EDW14676.1 KP071933 AJM71493.1 KX290639 APC94210.1 MH027968 AZB49386.1 GU575295 ADD70031.1 GDHF01001148 JAI51166.1 KU975055 AMY98994.1 MH937232 AYN70647.1 HG764550 CDJ79886.1 KQ460870 KPJ11697.1 GQ477025 ACZ58030.1 KT347569 ANA10230.1 HQ234489 ADY17884.1 HG764551 CDJ79887.1 KF460432 AHA39268.1 GAMC01003683 JAC02873.1 CH933809 EDW18016.1 KT381502 ALR72508.1 CH940656 EDW58548.1 MG546557 AYD42180.1 KP453838 ALM64969.1
Proteomes
Pfam
PF01395 PBP_GOBP
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
H9IZE5
A0A1E1W8T4
A0A345BER8
A0A2K8GKM7
A0A2K8GKL5
A0A2R4SBD9
+ More
A0A1B3P5J6 M9YYA4 A0A2K8GKN7 A0A0K8TUV6 C0SQ81 A0A0L7L7A8 A0A2K8GKQ9 H9IZE4 A0A076E7E0 A0A2H4G2B1 A0A076ED46 A0A1I9HZL2 A0A2K8GKN1 D6WS46 A0A1C8K2S9 A0A2R3ZV81 A0A310SBA3 D6WM81 G4Y3X2 A0A0X8RDP5 A0A1U9W501 D6WM83 A0A0S3J2L4 A0A1B4ZBJ7 A0A346TI01 A0A142FH84 U5YQF6 A0A1B1FKI3 A0A0T6BA30 A0A109NPF0 Q8WRW0 A0A1L2BL94 A0A0X8U7T9 A0A1W6QYA6 A0A1I8PZY0 D2D0C4 A0A1J0KKD4 A0A194PVX5 A0A1J0KKY1 B4KAZ8 A0A0C5BAY9 A0A1J0KKF9 A0A3G6V7G8 D4P5A8 A0A1I8N7H7 A0A0K8WJ37 A0A165UPF4 A0A3G2LEH8 W8W3V2 A0A194R1X8 F4Y5P3 A0A166IG76 F1CEY0 W8W3U3 A0A1S4FXS6 A0A075CKZ3 W8BUI1 B4L070 A0A0S3J2I5 B4MBS7 A0A386H995 A0A0X9EZ19
A0A1B3P5J6 M9YYA4 A0A2K8GKN7 A0A0K8TUV6 C0SQ81 A0A0L7L7A8 A0A2K8GKQ9 H9IZE4 A0A076E7E0 A0A2H4G2B1 A0A076ED46 A0A1I9HZL2 A0A2K8GKN1 D6WS46 A0A1C8K2S9 A0A2R3ZV81 A0A310SBA3 D6WM81 G4Y3X2 A0A0X8RDP5 A0A1U9W501 D6WM83 A0A0S3J2L4 A0A1B4ZBJ7 A0A346TI01 A0A142FH84 U5YQF6 A0A1B1FKI3 A0A0T6BA30 A0A109NPF0 Q8WRW0 A0A1L2BL94 A0A0X8U7T9 A0A1W6QYA6 A0A1I8PZY0 D2D0C4 A0A1J0KKD4 A0A194PVX5 A0A1J0KKY1 B4KAZ8 A0A0C5BAY9 A0A1J0KKF9 A0A3G6V7G8 D4P5A8 A0A1I8N7H7 A0A0K8WJ37 A0A165UPF4 A0A3G2LEH8 W8W3V2 A0A194R1X8 F4Y5P3 A0A166IG76 F1CEY0 W8W3U3 A0A1S4FXS6 A0A075CKZ3 W8BUI1 B4L070 A0A0S3J2I5 B4MBS7 A0A386H995 A0A0X9EZ19
PDB
6HHE
E-value=1.11014e-07,
Score=128
Ontologies
GO
Topology
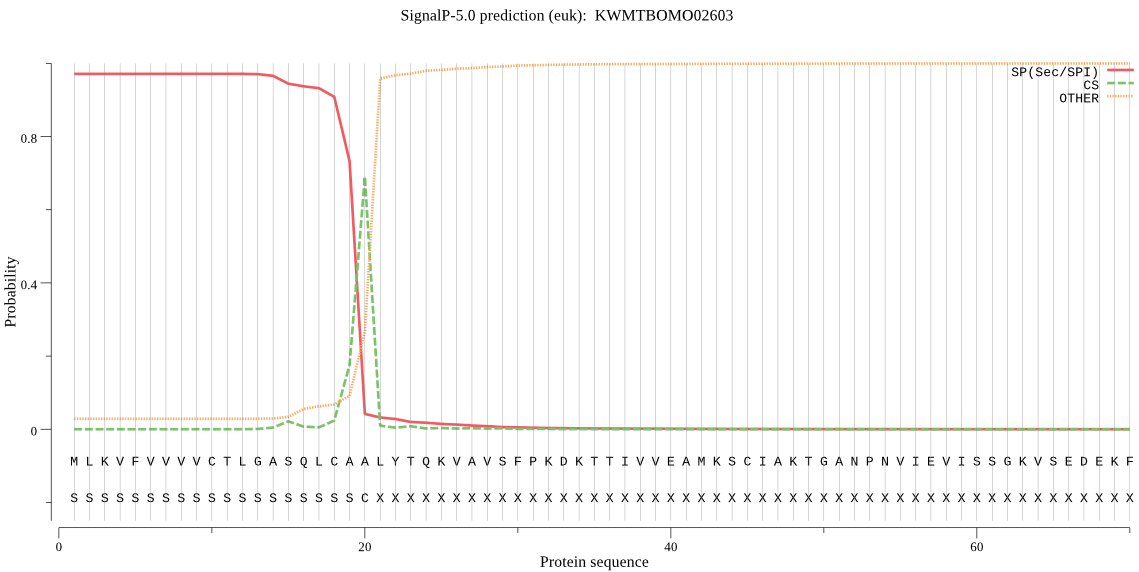
SignalP
Position: 1 - 20,
Likelihood: 0.971027
Length:
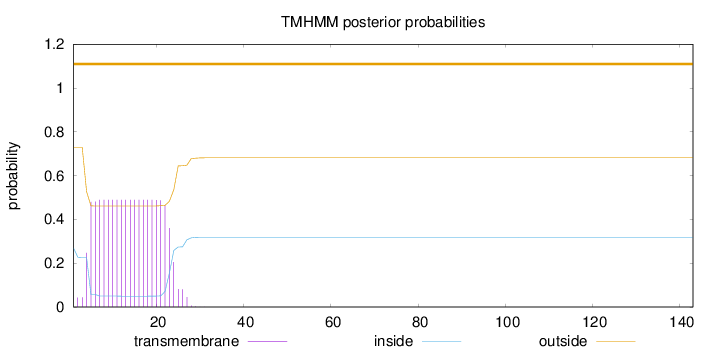
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.87793
Exp number, first 60 AAs:
9.87792
Total prob of N-in:
0.26990
outside
1 - 143
Population Genetic Test Statistics
Pi
160.21469
Theta
58.00846
Tajima's D
0.518077
CLR
0.210151
CSRT
0.51992400379981
Interpretation
Uncertain
